Session Information
Session Type: Poster Session B
Session Time: 9:00AM-10:30AM
Background/Purpose: The synovium encapsulates joints and contains fibroblasts which proliferate, become invasive, and drive disease progression in rheumatoid arthritis (RA) and osteoarthritis (OA). There is little known regarding normal synovial fibroblasts during homeostasis. Here, we have performed single cell RNA sequencing on healthy synovial tissue from humans.
Methods: Single cell RNA sequencing was performed on synovium from 16 human donors with no diagnosed arthritis, and on adipose tissue from 5 donors. The Bligh and Dyer method was used to fractionate fat conditioned media for lipidomic analysis. Bulk RNA sequencing was performed on cultured synovial fibroblasts. CRISPR-Cas9 was used to delete NR3C1 in synovial fibroblasts. In vitro 2D culture, and 3D synovial organoid culture, were implemented to assess cortisol anti-inflammatory and anti-fibrotic action. Readouts include immunofluorescence, qPCR, and flow cytometry.
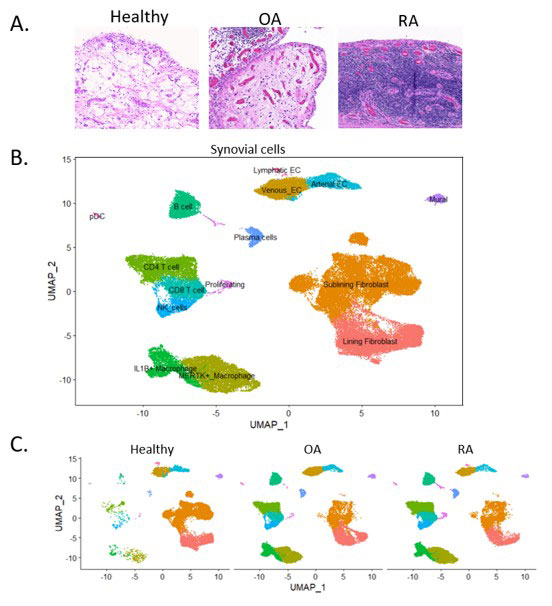
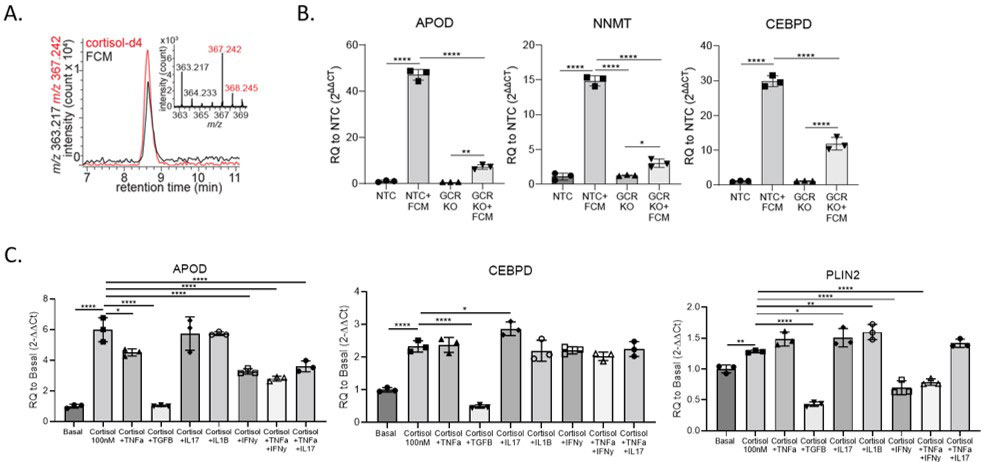
Results: Single cell RNA sequencing revealed that healthy synovium is primarily comprised of stromal cells (Fig. 1a-c). We identified a gene expression program which is enriched in homeostatic fibroblasts and characterized by enhanced fatty acid metabolism and lipid transport. The genes APOD, NNMT, and CEBPD, which are related to lipid metabolism were highly expressed in this gene expression program. Since normal synovial fibroblasts are surrounded by adipocytes, we stimulated fibroblasts with fat conditioned media (FCM) and used bulk RNA sequencing to find that the FCM gene signature is significantly enriched in healthy synovial fibroblasts. Through fractionation and lipidomic analysis of fat conditioned media, and validation using NRC31 (the glucocorticoid receptor gene) knockout cells, we discovered that cortisol is the primary factor driving this gene program. (Fig. 2a-b). We performed additional single cell RNA sequencing of adipose tissue fibroblasts. Using the Symphony package to map synovial fibroblasts onto adipose defined fibroblasts, we demonstrated strong mapping to committed preadipocytes. Additionally, committed preadipocytes are enriched in cortisol and FCM gene signatures compared to less committed DPP4+ universal fibroblast progenitors. We further defined the functional roles of cortisol in synovial fibroblast biology. Cortisol suppressed TNFα-induced MMP expression as well as TGFβ-induced fibrotic collagen expression. We found that IFNγ and TGFβ signaling is enhanced in RA and OA fibroblasts compared to normal fibroblasts, and these cytokines dampen cortisol signaling, potentially explaining why cortisol signaling is blunted in arthritis (Fig. 2c).
Conclusion: In sum, healthy synovium is characterized by high cortisol signaling which is also common to adipose tissue fibroblasts and is diminished in OA and RA fibroblasts.
To cite this abstract in AMA style:
Faust H, Cheng T, Korsunsky I, Watts G, Moody D, Brenner M. Molecular Profiling of Normal Human Synovium Reveals Striking Impact of Adipocytes and Homeostatic Cortisol Signaling [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/molecular-profiling-of-normal-human-synovium-reveals-striking-impact-of-adipocytes-and-homeostatic-cortisol-signaling/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/molecular-profiling-of-normal-human-synovium-reveals-striking-impact-of-adipocytes-and-homeostatic-cortisol-signaling/