Session Information
Date: Wednesday, October 29, 2025
Title: Abstracts: Rheumatoid Arthritis – Treatment II: Phenotyping and Personalization (2639–2644)
Session Type: Abstract Session
Session Time: 10:30AM-10:45AM
Background/Purpose: Rheumatoid arthritis (RA) is a common systemic autoimmune disease that targets the joints, causing persistent pain and swelling. Oral methotrexate (MTX) remains first-line therapy and an anchor drug for RA. However, up to 50% of patients do not achieve a clinically adequate outcome, and there is a lack of prognostic tools to predict MTX response. Variations in clinical outcomes may stem from interindividual differences in the gut microbiome. We and others have found differences in the pre-treatment microbiome of MTX responders (MTX-R) and non-responders (MTX-NR). However, whether there are consistent microbiome features associated with response in multiple cohorts remains unknown. Identification of cross-cohort features would enable development of a generalizable predictive model. Here, we investigate the microbiomes of 3 diverse patient cohorts and their associations with MTX response.
Methods: Three metagenomic datasets comprising fecal samples from 86 treatment-naïve RA subjects were analyzed. Two cohorts (Cohort 1, n=32; Cohort 2, n=25) were recruited from rheumatology clinics at New York University and University of California, San Francisco. Data for a third cohort (Cohort 3, n=29) was publicly available from Sequence Read Archive. Participants were new-onset RA patients. MTX response was defined as an improvement of ≥1.8 in the DAS28 at 12 to 16 weeks post-MTX monotherapy. MMUPHin was employed for metagenomic batch correction and data harmonization. Microbiome differences were assessed using PERMANOVA.
Results: After batch correction, when examining all three cohorts, we confirmed that the pretreatment gut microbiome community composition differed between MTX-R and MTX-NR patients. Of 2,386 strains observed, 95 were differentially abundant (punadj < 0.05), with 24 increased and 71 decreased in MTX-NR. Key gut commensal strains were decreased in MTX-NR, including multiple strains from the Faecalibacterium, Clostridium, and Bacteroides genera. Functional profiling of the microbiome identified 8,895 KEGG Orthologs (KOs), of which 252 were associated with response. Intriguingly, we observed decreased abundance of 231 KOs in MTX-NR that are likely direct and indirect targets of MTX, including dihydrofolate reductase (K00287), methylenetetrahydrofolate dehydrogenase (K01491), and genes associated with metabolism of pyrimidines (K01591), purines (K01939), and histidine (K00013) (punadj< 0.05). Of 601 pathways examined, 19 differed by MTX response status. Biosynthesis pathways for L-histidine, L-tyrosine, L-lysine, and inosine-5'-phosphate and the superpathway for pyrimidine salvage were reduced in MTX-NR (punadj < 0.05). Finally, we applied machine learning techniques to the metagenomic data, resulting in a model that predicted response to MTX.
Conclusion: Our study confirms that the pretreatment gut microbiome is associated with response to MTX in new-onset RA across multiple diverse cohorts. We identify multiple taxa, microbial functions, and pathways that are reproducibly associated with clinical response in patients. Development of a robust predictive model will bring us closer to the goal of advancing precision medicine for patients with RA.
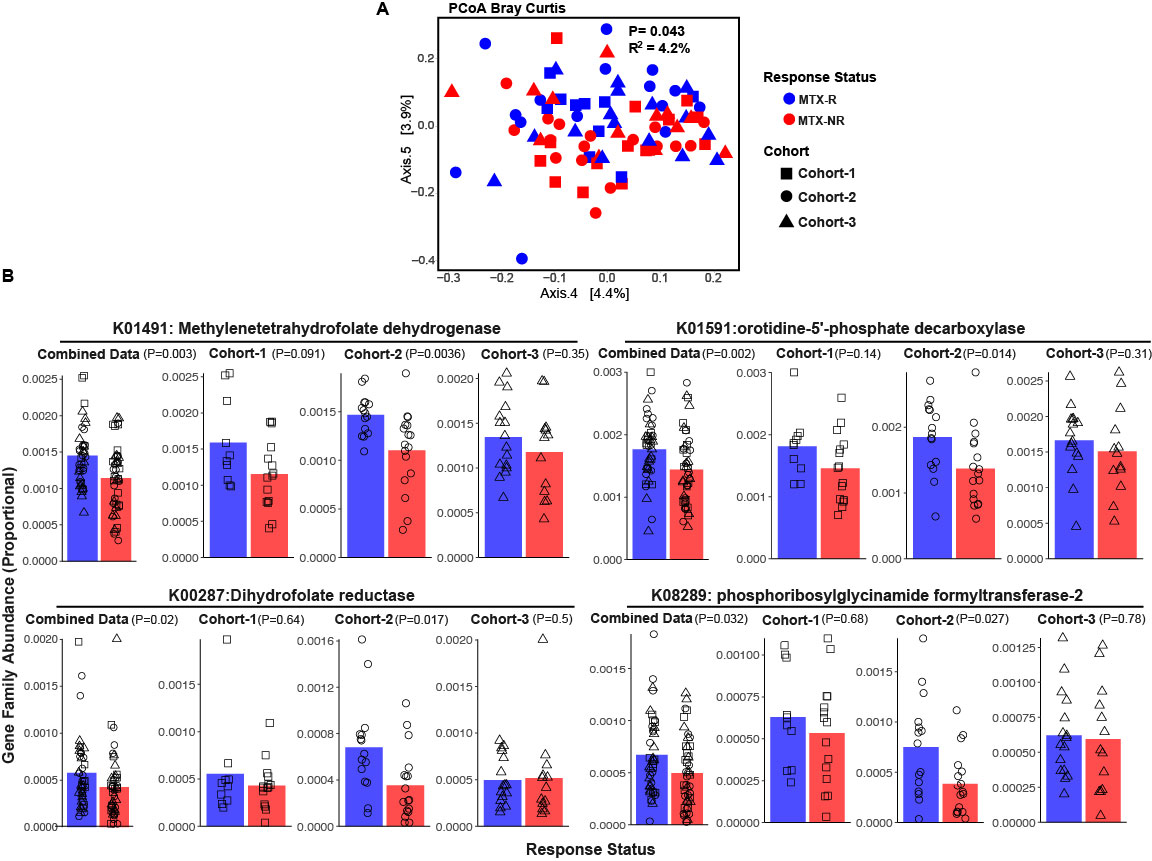 Figure 1. Pretreatment gut microbiome differs between methotrexate responders (MTX-R) and non-responders (MTX-NR) in patients with new-onset rheumatoid arthritis (RA). A. Principal coordinates analysis (PCOA) of pretreatment microbiota composition from MTX-R and MTX-NR groups, based on Bray-Curtis distances (p=0.046, R2 = 0.042). B. Differentially abundant KEGG orthologs that are likely to be direct or indirect targets of MTX identified using a mixed-effects model on combined data and Wilcoxon tests within individual cohorts.
Figure 1. Pretreatment gut microbiome differs between methotrexate responders (MTX-R) and non-responders (MTX-NR) in patients with new-onset rheumatoid arthritis (RA). A. Principal coordinates analysis (PCOA) of pretreatment microbiota composition from MTX-R and MTX-NR groups, based on Bray-Curtis distances (p=0.046, R2 = 0.042). B. Differentially abundant KEGG orthologs that are likely to be direct or indirect targets of MTX identified using a mixed-effects model on combined data and Wilcoxon tests within individual cohorts.
To cite this abstract in AMA style:
Bodkhe R, Trepka K, Orellana D, Blank R, Turnbaugh P, Scher J, Nayak R. Microbiome Signatures in RA Treatment: Personalizing Methotrexate Therapy [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/microbiome-signatures-in-ra-treatment-personalizing-methotrexate-therapy/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/microbiome-signatures-in-ra-treatment-personalizing-methotrexate-therapy/
