Session Information
Session Type: Abstract Session
Session Time: 4:00PM-5:30PM
Background/Purpose: Osteoarthritis (OA) is a diverse and multifaceted condition. Despite its widespread incidence, the core mechanisms of the disease remain elusive, and therapeutic interventions are restricted. The complexity of OA is compounded by the absence of well-defined subgroups or “endotypes” that could elucidate specific triggers and guide towards more personalized treatments. Recent studies propose that molecular biomarkers could distinguish various OA endotypes. For instance, certain biomarkers might show elevated levels in OA patients experiencing inflammation, while others might increase in those with advanced cartilage deterioration.
This study aimed to uncover OA endotypes via soluble biomarkers of tissue turnover, using unsupervised machine learning methodologies.
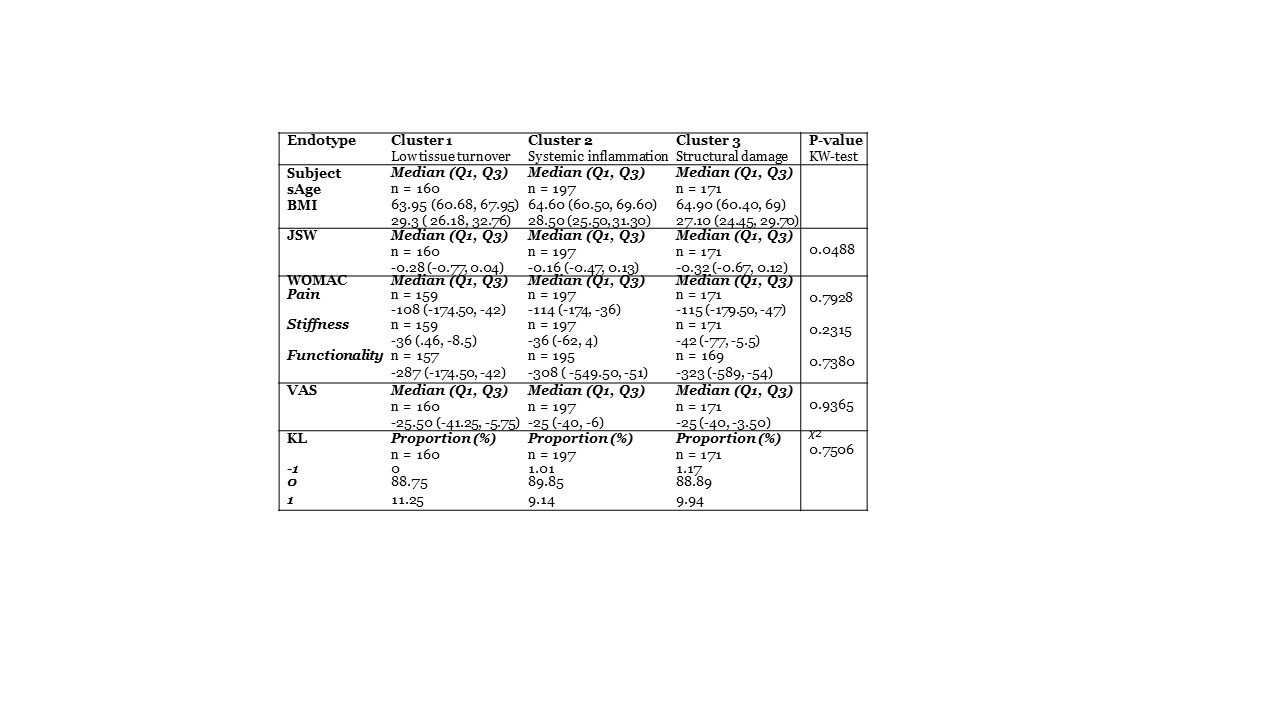
Methods: The study measured biomarkers of cartilage alteration (CTX-II, C2M, T2CM, PRO-C2), bone modification (N-MID, UCTX-I, SCTX-I), and tissue inflammation (CRPM, VICM, C1M, C3M) were measured at baseline in phase III clinical trials SMC01 (n=1176) and SMC02 (n=1030) which tested the effectiveness and safety of oral salmon calcitonin in knee OA patients. The study included only placebo group patients who had more than 5 biomarkers available (n=528). About 4% of data was missing and was imputed using Random Forest. K-Means clustering was employed on UMAP dimensionality-reduced data. Associations between the changes in symptomatic and radiographic parameters over two years with the identified clusters were compared using the Kruskal Wallis test for numerical variables and chi-square test for categorical variables.
Results: The K-Means clustering resulted in three unique clusters; 1) a low tissue turnover endotype, 2) a systemic inflammation endotype and 3) a structural damage endotype (figure 1). The structural damage endotype showed elevated levels of bone remodeling markers CTX-I, N-MID, and the cartilage degradation marker CTX-II. The systemic inflammation endotype was characterized by higher levels of C1M, C2M, C3M, CRPM, and VICM associated with tissue inflammation. Conversely, the low tissue turnover endotype displayed generally lower levels of both tissue-related and inflammatory markers. A difference in JSW change over two years was noted among the three groups (Cluster 1: -0.28 mm, Cluster 2: -0.16 mm, Cluster 3: -0.32 mm; p=0.049), but the difference was not statistically significant in the adjusted analysis. Broadly, the three endotypes showed similar progression.
Conclusion: These findings indicate the presence of unique biomarker-based endotypes in OA, underlining the fact that several mechanistic pathways may result in joint degradation. Identifying these endotypes could facilitate the development of more precise treatments that are designed to address specific causes of OA in distinct patient subgroups.
To cite this abstract in AMA style:
Hannani M, Lisowska-Petersen Z, Karsdal M, Bager C, Bay-Jensen A, Thudium C. From Biomarkers to Endotypes: Data-driven Identification of Knee Osteoarthritis Patients Subtypes [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/from-biomarkers-to-endotypes-data-driven-identification-of-knee-osteoarthritis-patients-subtypes/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/from-biomarkers-to-endotypes-data-driven-identification-of-knee-osteoarthritis-patients-subtypes/