Session Information
Date: Monday, November 18, 2024
Title: Plenary III
Session Type: Plenary Session
Session Time: 9:00AM-10:30AM
Background/Purpose: In rheumatoid arthritis (RA), synovial fibroblasts adopt an inflammatory phenotype that catalyzes pathogenic synovial hyperplasia and ultimately bone and cartilage destruction. However, identifying therapeutic entry points to modulate synovial fibroblast phenotypes remains challenging. Here, we describe the development and execution of an in vitro arrayed CRISPR deletion screen and identify novel molecular regulators of stromal inflammation.
Methods: To generate a custom arrayed CRISPR library for screening, integrated analyses of bulk and single-cell RNA-sequencing (scRNAseq) datasets from OA and RA were used to generate a composite ranking of all genes based on several criteria including expression, upregulation in RA, enrichment in inflammatory fibroblast subsets, fibroblast specificity, and predicted functional relevance. For CRISPR deletion, synovial fibroblasts were nucleofected with guide RNA (gRNA)-Cas9 ribonucleoprotein complexes targeting gene candidates and controls. Cells were cultured for 7 days and stimulated for 16 hours with a combination of TNF-α, IL-17, IFN-γ, and IL-1β, cytokines relevant in RA pathogenesis. Cells and supernatant were processed for downstream bulk RNAseq and ELISA analyses, Western blot, qPCR, and functional assays. Genes upregulated on cytokine stimulation of non-transfected controls were used to develop stromal inflammation signatures.
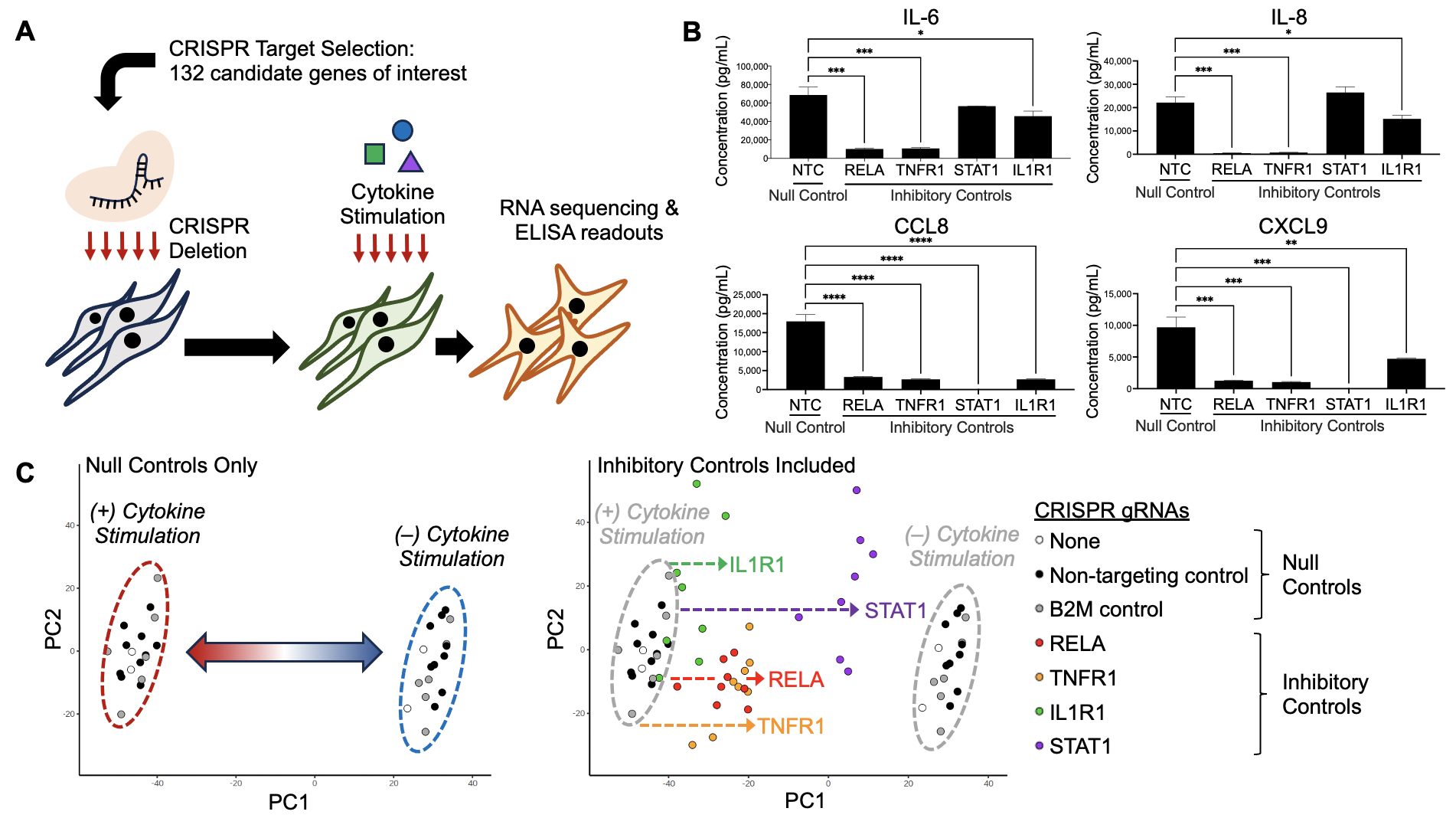
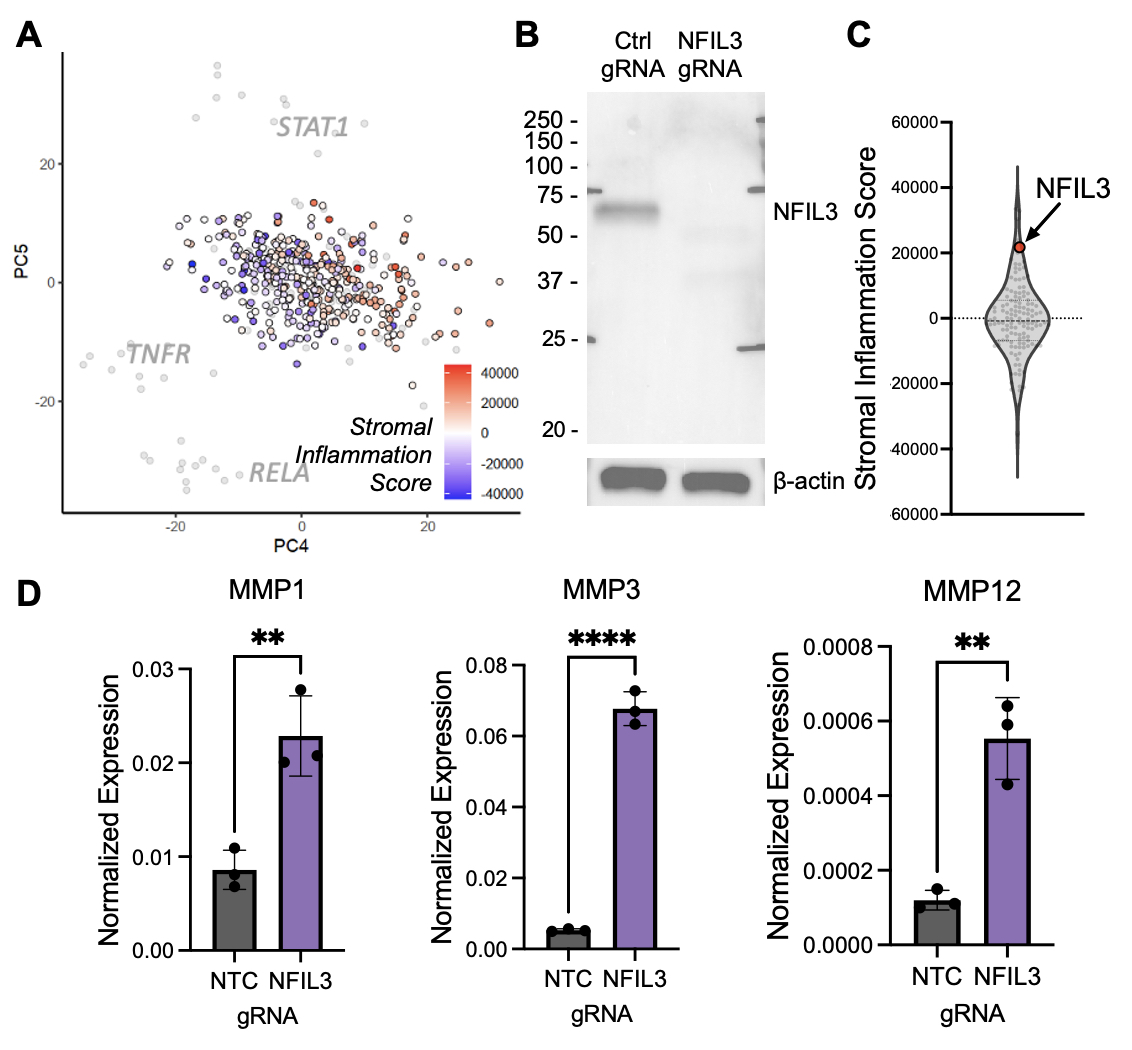
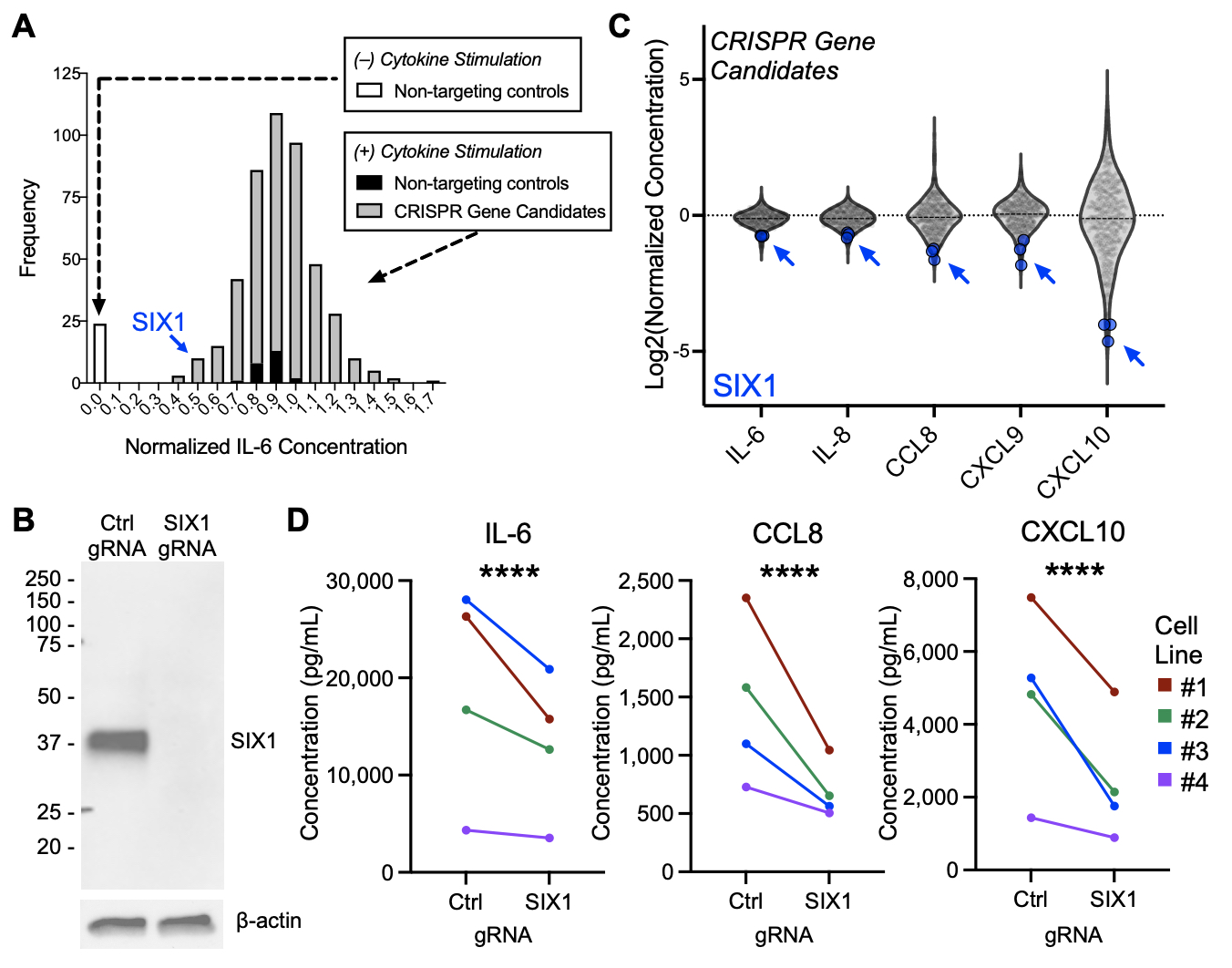
Results: We performed individual CRISPR deletion of 132 genes in an arrayed screen of human RA synovial fibroblasts and evaluated the impact on inflammatory cytokine activation through RNAseq of cell lysates and ELISA of cell culture supernatant (Fig 1A). As a proof-of-concept, we show that deletion of genes known to be active in RA signaling pathways, including RELA, TNFR1, STAT1, and IL1R1, abrogates secretion of pathogenic cytokines (Fig 1B) and attenuates transcriptional changes associated with cytokine stimulation (Fig 1C). Two methods were used for hit calling: one based on enrichment in a stromal inflammation gene signature (Fig 2) and another based on protein expression (Fig 3), and we highlight one hit from each approach. In RNAseq analyses, stromal inflammation signature scores were calculated to identify genes whose CRISPR deletion enhanced or decreased inflammatory potentiation (Fig 2A). Targeting NFIL3 (Fig 2B), a transcription factor implicated in JIA and arthritis severity in mouse models, results in strong enrichment of the inflammation gene signature (Fig 2C) and upregulation of matrix metalloproteases (Fig 2D), suggestive of an invasive phenotype. In protein analyses, the concentrations of ten selected inflammation markers were measured by ELISA to identify candidates that altered cytokine and chemokine secretion (Fig 3A, IL-6 shown). Deletion of SIX1 (Fig 3B), a transcription factor known to promote cell proliferation and tumorigenesis, potently decreased secretion of several cytokines (Fig 3C), which was validated in multiple RA cell lines (Fig 3D).
Conclusion: Here, we show the utility of CRISPR deletion screens to identify novel regulators of fibroblast activation and pathologic effector functions. Hits identified and pathways they regulate may be considered for drug development.
To cite this abstract in AMA style:
Mueller A, Kongthong S, Zou A, Watts G, Murphy C, Nguyen H, Perrigoue J, Chamberlain M, Wei K, Raychaudhuri S, Brenner M. CRISPR Deletion Screen in Fibroblasts Identifies Novel Regulators of Inflammation [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/crispr-deletion-screen-in-fibroblasts-identifies-novel-regulators-of-inflammation/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/crispr-deletion-screen-in-fibroblasts-identifies-novel-regulators-of-inflammation/