Session Information
Date: Tuesday, October 28, 2025
Title: (2377–2436) Systemic Lupus Erythematosus – Diagnosis, Manifestations, & Outcomes Poster III
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: During disease, ~50% of SLE patients develop lupus nephritis (LN), one of the most serious complication. Despite the best therapy, within 15 years, ~20% progress to renal failure (1). Diagnosis is based on urine protein-creatinine ratio and histology in renal biopsy; genetic factors are common, but penetrance is low, and the cause is unknown. A gut-kidney axis has been suggested. We therefore investigated pathogenetic urinary factors associated with host response to an implicated pathobiont in biopsy-proven patients.
Methods: Patients with SLE diagnosis met ACR criteria and had a SLE Disease activity scores. Urine protein-creatinine ratio was determined at the time of sample collection. As a marker of microbe-specific immune exposure, serum antibodies to cell wall lipoglycan (LG) in pathogenic strains of Ruminococcus gnavus (RG) were quantified by a standardized bead-based assay (2,3). Compared to active LN without dysbiosis, active LN with RG gut bloom and high anti-LG had elevated transcripts reflecting platelet (SPARC, SNCA, PROS1, PF4, PDGFA, MMRN1, EGF) and neutrophil activation (GATA1)(padj=2×10(-4))in whole blood(manuscript submitted). We therefore used the same proteomics assay as in AMP-sponsored high-dimensional discovery studies to detect self-proteins in urine to identify WHO class and activity of glomerular lesions by urine “liquid biopsy” (4). In contrast, we custom-designed multiplex arrays to detect 30 human proteins implicated in platelet-associated thrombo-inflammation and neutrophil extracellular TRAPs/NETosis, and in other immune pathways. By urine hybridization, antigens were detected, data normalized by creatinine levels.
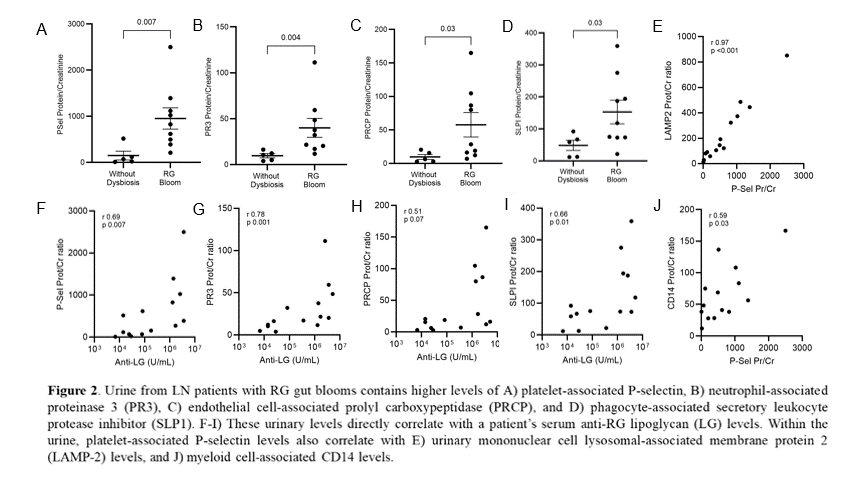
Results: With this custom proteomics array, we tested urine of 14 LN at flare with coincident RG blooms and 6 LN patients at flare without bloom. For selected analytes (normalized to urine creatinine), significant increases were seen in urine of patients with gut dysbiosis (Figs. 1A-D). Analytes were proteins from platelets, neutrophils and endothelial cells. These data support the hypothesis that platelet & neutrophil-related pathways contribute to LN activity during RG blooms (Figs. 1A, B). Presence of these proteins in urine is positively associated with RG blooms, with significant correlations of platelet and neutrophil proteins with serum antibodies to RG lipoglycan (Figs. 1F, G). Findings were highly reproducible.Hence, renal exposure to either the lipoglycan, or the induced antibodies to lipoglycan, may contribute to glomerular injury.
Conclusion: Our studies suggest that LN flares in patients with gut RG blooms are associated with peripheral platelet activation similar to that observed in patients with sepsis. In the affected LN patients, the level of urinary proteins from activated platelets, neutrophils, and endothelial cells documents underlying systemic immune activation, with release of proteases that likely injure the kidney. Taken together, these data indicate that intestinal expansions of the RG pathobiont can worsen LN flares through thrombo-inflammation. This previously unsuspected endotype of LN may be amenable to therapeutic interventions beyond the glucocorticoids and immunosuppressants commonly used to treat LN.
To cite this abstract in AMA style:
Silverman G, Amarnani A, Azad Z, Rovin B. Identification of an LN endotype linked to intestinal expansion of Pathogenic Strains of a Pathobiont Bacterium that induces Systemic Thrombo-inflammatory Pathways directly measurable in Urine [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/identification-of-an-ln-endotype-linked-to-intestinal-expansion-of-pathogenic-strains-of-a-pathobiont-bacterium-that-induces-systemic-thrombo-inflammatory-pathways-directly-measurable-in-urine/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-an-ln-endotype-linked-to-intestinal-expansion-of-pathogenic-strains-of-a-pathobiont-bacterium-that-induces-systemic-thrombo-inflammatory-pathways-directly-measurable-in-urine/