Session Information
Date: Tuesday, October 28, 2025
Title: Plenary III (1722–1727)
Session Type: Plenary Session
Session Time: 8:30AM-8:45AM
Background/Purpose: Mucosal-associated invariant T (MAIT) cells are innate-like lymphocytes that produce cytokines and cytotoxic molecules. Reduced frequencies and altered phenotypes of MAIT cells have been described in autoimmune diseases. Their ability to interact with microbiota and accumulate in inflamed tissues suggests a role in RA pathogenesis.The objective was to explore the phenotypic, functional, and transcriptomic alterations of MAIT cells in blood and synovial fluid (SF) in RA and assess their impact on inflammation and joint damage using in vitro and in vivo models.
Methods: MAIT cells were analyzed by flow cytometry in peripheral blood from 75 RA patients and 42 healthy donors (HD). SF MAIT cells were studied in 19 RA patients. Single-cell RNA sequencing (scRNA-seq) was performed on paired blood and SF samples from 6 RA patients and 4 HDs. MAIT–fibroblast-like synoviocyte (FLS) interactions were assessed in co-culture. To study their in vivo role, MAIT cells were modulated in two arthritis mouse models—mBSA (methylated bovin serum albumin)-induced and TNF-transgenic—using MR1 knockout mice and Ac-6-FP, a pharmacological inhibitor of MAIT cell activation.
Results: Circulating MAIT cells were reduced in RA compared to HD (0.51% vs 2.7%, p< 0.001), with an activated/exhausted phenotype (↑CD69, CD25, Tim-3) and increase production of IL-17 and granzyme B. scRNA-seq showed upregulation of genes linked to activation, exhaustion, apoptosis, cytotoxicity (GZMB, PRF1), and chemokines receptors (CCR1, CXCR6). Despite their depletion in blood, MAIT cells were enriched in SF (0.65% vs 0.32%, p=0.048), especially in early arthritis. SF MAIT cells were more activated (↑CD69, CD25), exhausted (↑PD-1, ↓CD127), and had lower CCR6 and CD56 expression. scRNA-seq confirmed this profile, and revealed type I IFN pathway engagement, as well as chemokine-driven migration signature. CellChat analysis implicated pDCs and monocytes in generating a chemokine gradient recruiting MAIT cells into joint. SF MAIT cells also showed increased IL-10, suggesting combined inflammatory and regulatory roles.Activated MAIT cells enhanced FLS activation in co-culture, increasing ICAM1 and PDPN expression, and secretion of IL-1β, IL-6, IL-8, and MCP-1. Bulk RNA-seq of FLS exposed to activated MAIT cells showed upregulation of inflammatory cytokines, chemokines, IFN-stimulated genes, and matrix-degrading enzymes.In vivo, MR1 KO mice showed reduced arthritis severity and joint damage in both models. Ac-6-FP treatment also improved clinical and histological outcomes in mBSA arthritis.
Conclusion: MAIT cells are reduced in blood but accumulate in RA synovial fluid, where they are highly activated and exhausted, likely shaped by local inflammation. Their migration from blood to the joint may be driven by a synovial chemokine gradient. In the joint, they promote inflammation through cytotoxicity, cytokine production, and FLS activation. Notably, their IL-10 expression suggests additional immunoregulatory functions. Modulating MAIT cells reduces arthritis in murine models, supporting their therapeutic potential in RA.
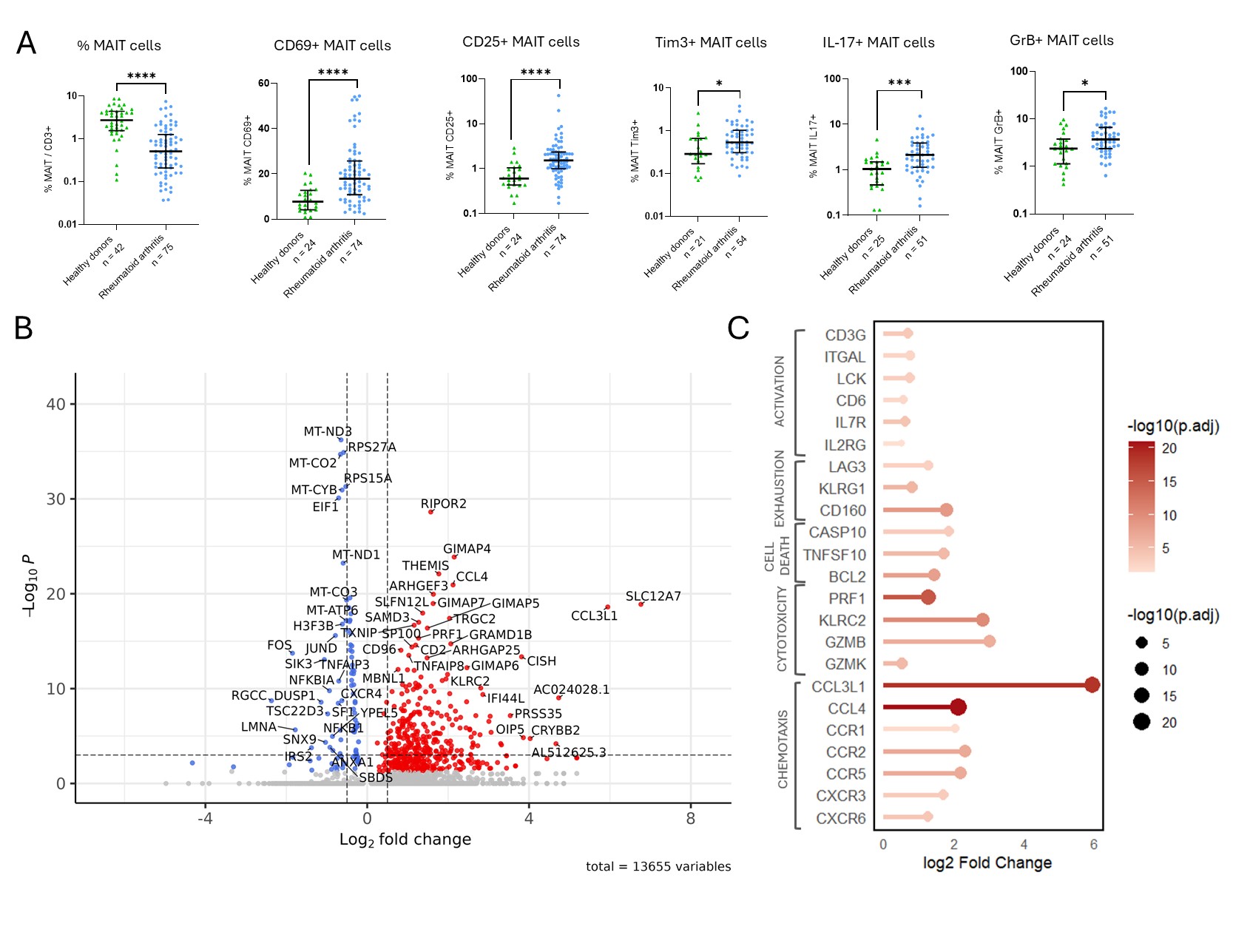 Figure 1. Phenotypic and transcriptomic alterations of circulating MAIT cells in RA.
Figure 1. Phenotypic and transcriptomic alterations of circulating MAIT cells in RA.
(A) Frequency and functional profile of circulating MAIT cells in RA patients compared to healthy donors (HD), including expression of CD69, CD25, Tim-3, IL-17, and granzyme B after stimulation. (B) Volcano plot showing differentially expressed genes in RA MAIT cells versus HD, based on single-cell RNA sequencing (scRNA-seq). (C) Lollipop plot summarizing selected upregulated genes in RA MAIT cells, categorized into functional groups: activation, exhaustion, apoptosis, cytotoxicity, and chemotaxis.
Statistical comparisons were performed using Mann–Whitney test.
MAIT: mucosal-associated invariant T cell; RA: rheumatoid arthritis; HD: healthy donor; scRNA-seq: single-cell RNA sequencing.
.jpg) Figure 2. Phenotypic and transcriptomic profile of MAIT cells in synovial fluid.
Figure 2. Phenotypic and transcriptomic profile of MAIT cells in synovial fluid.
(A) Frequency and surface marker expression of MAIT cells from synovial fluid (SF) of RA patients compared to paired peripheral blood MAIT cells and blood MAIT cells from healthy donors (HD), including CD69, CD25, PD-1, and CD56.
(B) UMAP projection of all MAIT cells analyzed by single-cell RNA sequencing (scRNA-seq), colored by sample origin: HD blood, RA blood, and RA SF.
(C) Lollipop plot showing selected differentially expressed genes in RA SF MAIT cells compared to RA blood MAIT cells, grouped by function: activation, activation/migration, migration, exhaustion, and interferon (IFN) pathway.
(D) Circos plot illustrating CellChat-predicted interactions between chemokine receptors expressed by circulating MAIT cells (e.g., CXCR3, CXCR6) and their ligands expressed by synovial immune cell subsets.
Statistical comparisons were performed using Wilcoxon Test.
MAIT: mucosal-associated invariant T cell; RA: rheumatoid arthritis; SF: synovial fluid; HD: healthy donor; scRNA-seq: single-cell RNA sequencing; IFN: interferon.
.jpg) Figure 3. In vivo modulation of MAIT cells in two murine models of arthritis.
Figure 3. In vivo modulation of MAIT cells in two murine models of arthritis.
(A) Clinical arthritis score and histological inflammation score in the mBSA-induced arthritis model, comparing MR1 knockout (KO) mice (MAIT cell-deficient) to wild-type controls.
(B) Effect of pharmacological MAIT cell inhibition using Ac-6-FP versus PBS control in the mBSA model: clinical score and histological inflammation score.
TNF arthritis in MR1 KO mice compared to controls :
(C) Micro-computed tomography (microCT) assessment of bone erosions showing increased bone volume fraction (BV/TV) in MR1 KO mice compared to controls.
(D) Histological inflammation score in TNF-transgenic MR1 KO and control mice.
Statistical comparisons were performed using Student T test.
MAIT: mucosal-associated invariant T cell; MR1 KO: MR1 knockout; mBSA: methylated bovine serum albumin; BV/TV: bone volume/total volume; PBS: phosphate-buffered saline.
To cite this abstract in AMA style:
LESTURGIE-TALAREK M, Gonzalez V, Cauvet A, Beaudoin L, Oudart F, Schvartz A, Carbone F, Menager M, Sénot N, Tilotta F, Allanore Y, Lehuen A, AVOUAC J. Pathogenic role of Mucosal-Associated Invariant T cells in Rheumatoid Arthritis. [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/pathogenic-role-of-mucosal-associated-invariant-t-cells-in-rheumatoid-arthritis/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/pathogenic-role-of-mucosal-associated-invariant-t-cells-in-rheumatoid-arthritis/
