Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: The etiology of rheumatoid arthritis (RA) is not entirely known. Epigenetic modifications could be the link between genetic and environmental factors related to the appearance and evolution of the disease. RA patients present dysbiosis in different grades, and a lower microbiota diversity compared to healthy individuals. To date, literature regarding the relation between microbiota and epigenetic modifications is scarce.
Our objective in this study was to analyse the relationship between gut microbiota, inflammation, and epigenetics modification in RA patients .
Methods: Cross-sectional study in a prospective cohort of 110 RA patients (ACR/EULAR 2010 criteria) and 110 healthy controls without inflammatory condition, matched by age and sex. Gut microbiota was evaluated by NGS platform Ion Torrent S5 and the obtained sequences were processed by Quantitative Insights Into Microbial Ecology 2 software. DNA methylation was determined by Infinium Methylation EPIC (HM850K) BeadChip (Illumina, San Diego, CA, USA), cytosine methylation level was expressed as β-value. The validation of the results of HM850K was performed by pyrosequencing, on PyroMark Q48 (Qiagen, Hilden, Germany), including data about CpGs bordering CpGs of the array. The other variables included were epidemiological, clinical-analytical, and mean DAS28-ESR from RA onset to the cross-sectional visit. Descriptive analysis, bivariant analysis, t-student, χ2, and multivariate analysis were performed.
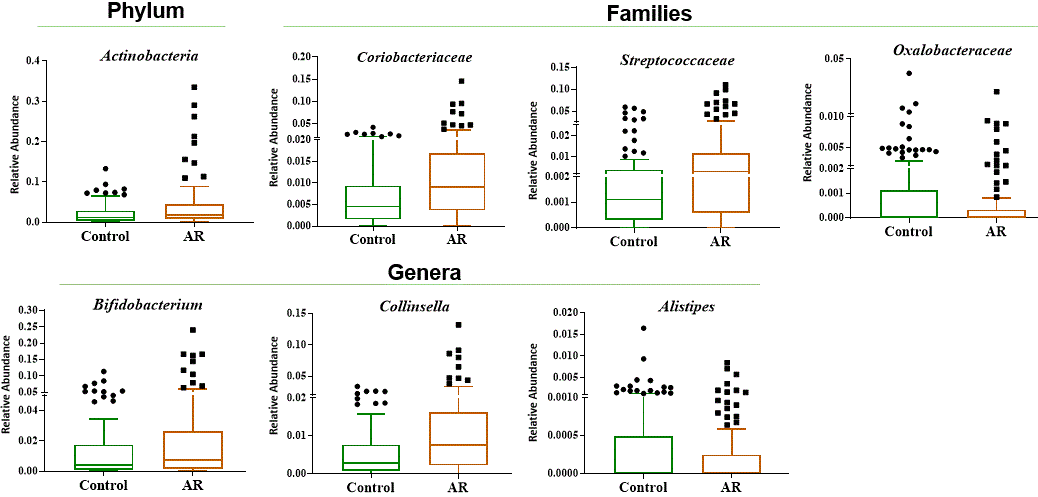
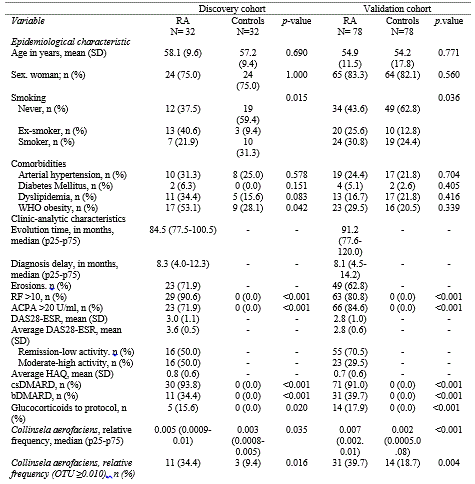
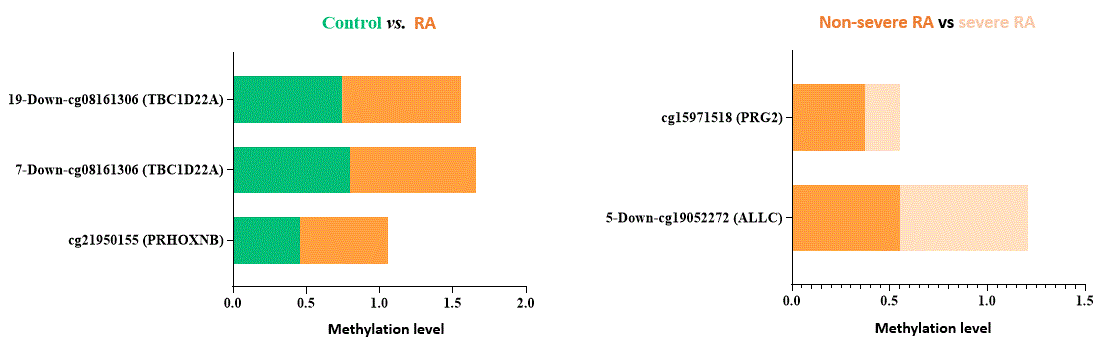
Results: Table 1 shows that RA patients’ group had more smokers (p=0.015), higher frequency of obesity (p=0.042), and higher abundance of Collinsella aerofaciens (p=0.047). Logistic regression analysis evidenced that elevated CpG methylation in PRHOXNB, TBC1D22A and PRG2, which cover specific sites such as cg21950155, cg08161306 and cg15971518, were associated with RA diagnosis, as well as decreased methylation in ALLC (cg19052272). RA patients with moderate/high activity (DAS28≥3.2) displayed lower methylation levels in cg19052272 site (ALLC) (75.7± [13.1] vs 87.7 [5.4]; p=0.003) compared to remission/low activity patients. Collinsella showed a positive correlation with cg08161306 (TBC1D22A) (r=0.267; p=0.033), and an inverse correlation with cg19052272 (ALLC) (r= -0.398; p=0.024). A linear regression model established that bacteria associated in RA to cg19052272 (ALLC) were Actinobacteria (β=-0.434; p=0.013), Coriobacteriaceae (β=-0.469; p=0.007), Collinsella (β=-0.398; p=0.024), Alistipes (β=-0.404; p=0.022) and Bifidobacterium (β=-0.384; p=0.030), which also negatively correlated with methylation levels.
Conclusion: Gut microbiota, particularly Collinsella, could be associated with both methylation patterns and inflammatory in RA patients, so they could be potential biomarkers for diagnosis and prognosis of this entity.
The graphs represent the taxonomic levels of phylum, families and genera with statistically significant differences between controls and RA patients.
ACPA: anti-citrullinated protein autoantibodies; RF: rheumatoid factor; SD: Standard deviation; DAS28-ESR: Disease Activity Score_28 for Rheumatoid Arthritis with ESR; ESR: Erythrocyte Sedimentation Rate; csDMARD: conventional synthetic Disease-modifying antirheumatic drug; bDMARD: biological Disease-modifying antirheumatic drug; WHO: World Health Organization.
CpGs related to genes or pseudogenes, β value >0.10 and p value ≤ 0.01.
To cite this abstract in AMA style:
Lisbona-Montañez j, Mucientes A, Ruiz-Limón P, Martín-Nuñez G, Redondo-Rodríguez R, Cano-García L, Manrique-Arija S, Moreno-Indias I, Mena Vázquez N, Fernández-Nebro A. Association Between Gut Microbiota, Inflammation, and Epigenetics in Rheumatoid Arthritis Patients [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/association-between-gut-microbiota-inflammation-and-epigenetics-in-rheumatoid-arthritis-patients/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/association-between-gut-microbiota-inflammation-and-epigenetics-in-rheumatoid-arthritis-patients/