Session Information
Date: Sunday, November 12, 2023
Title: Abstracts: Systemic Sclerosis & Related Disorders I: Translational
Session Type: Abstract Session
Session Time: 2:00PM-3:30PM
Background/Purpose: SSc-ILD is the leading cause of death in SSc affecting around 50% of the patients. Lung tissue of patients with early-stage SSc-ILD is characterized by a predominant inflammatory response with inconspicuous fibrosis, which can progress to honeycombing fibrosis. Although several biomarkers and drug targets have been proposed to monitor and halt SSc-ILD progression, none has reached clinical application. A better understanding of the molecular mechanisms underpinning SSc-ILD pathogenesis is needed to improve treatment options and progression prediction. This transcriptomic study aims to reveal the differential gene expression between healthy control (ctrl) lung sections and regions of interest (ROIs) within SSc-ILD lung tissue.
Methods: The nanoString (nS) nCounter Human Fibrosis Panel containing 770 genes related to all stages of fibrosis was used to analyze gene expression in formalin-fixed and paraffin-embedded lung tissues with varying stages of SSc-ILD (n=18) and control lung tissue (n=6). The SSc-ILD tissues were stratified by an experienced lung pathologist into three ROIs, inflammatory (inf), prefibrotic (prefib), or fibrotic (fib). Every group comprised 6 samples. This stratification aimed to define a longitudinal simulation of early to late phases of SSc-ILD: ctrl à inflammation à prefibrosis à fibrosis. nSolver and Rosalind software were used for data and statistical analysis. The immunohistochemistry was performed using specific antibodies in the same tissues used for nS analysis to confirm protein expression.
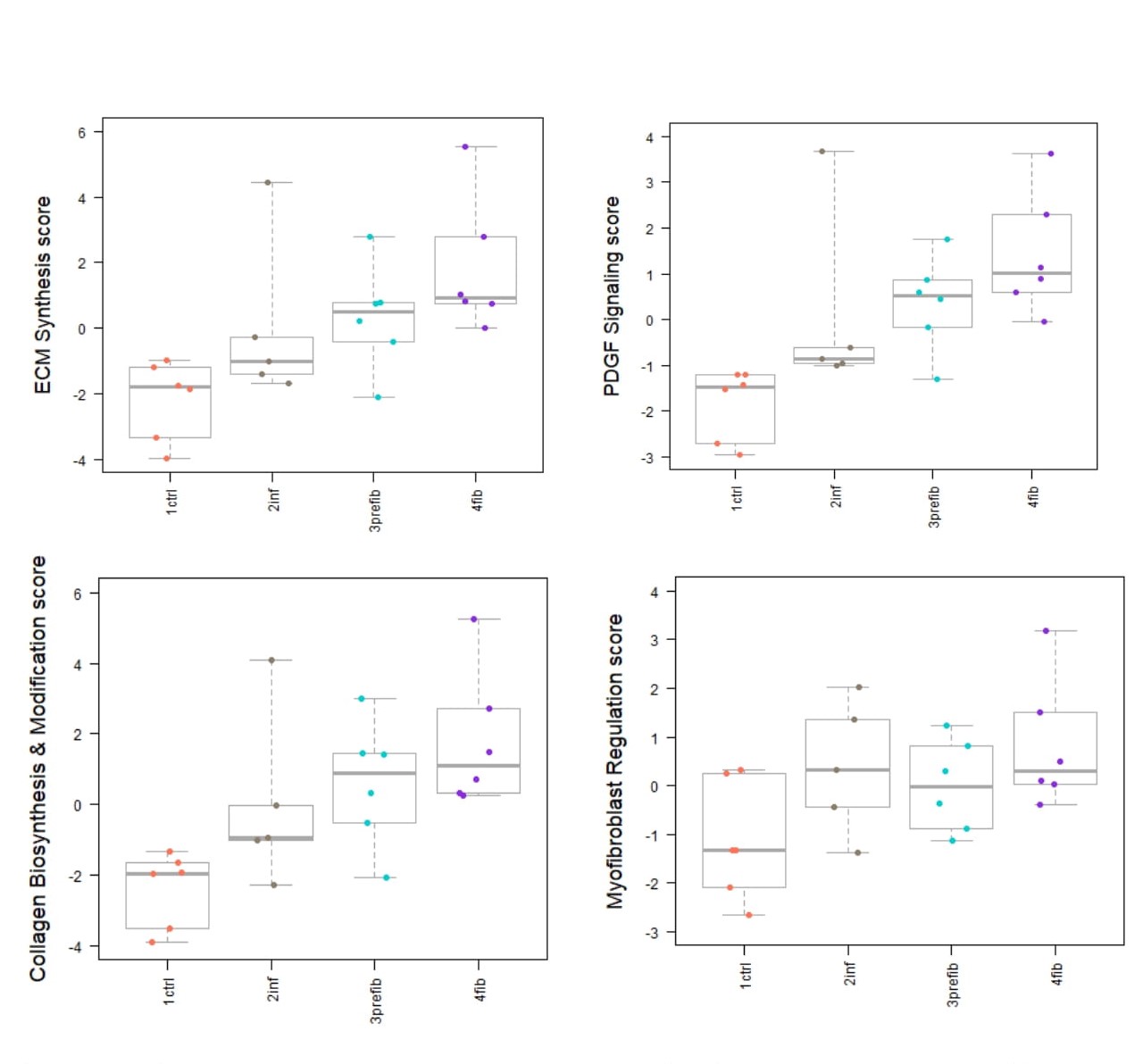
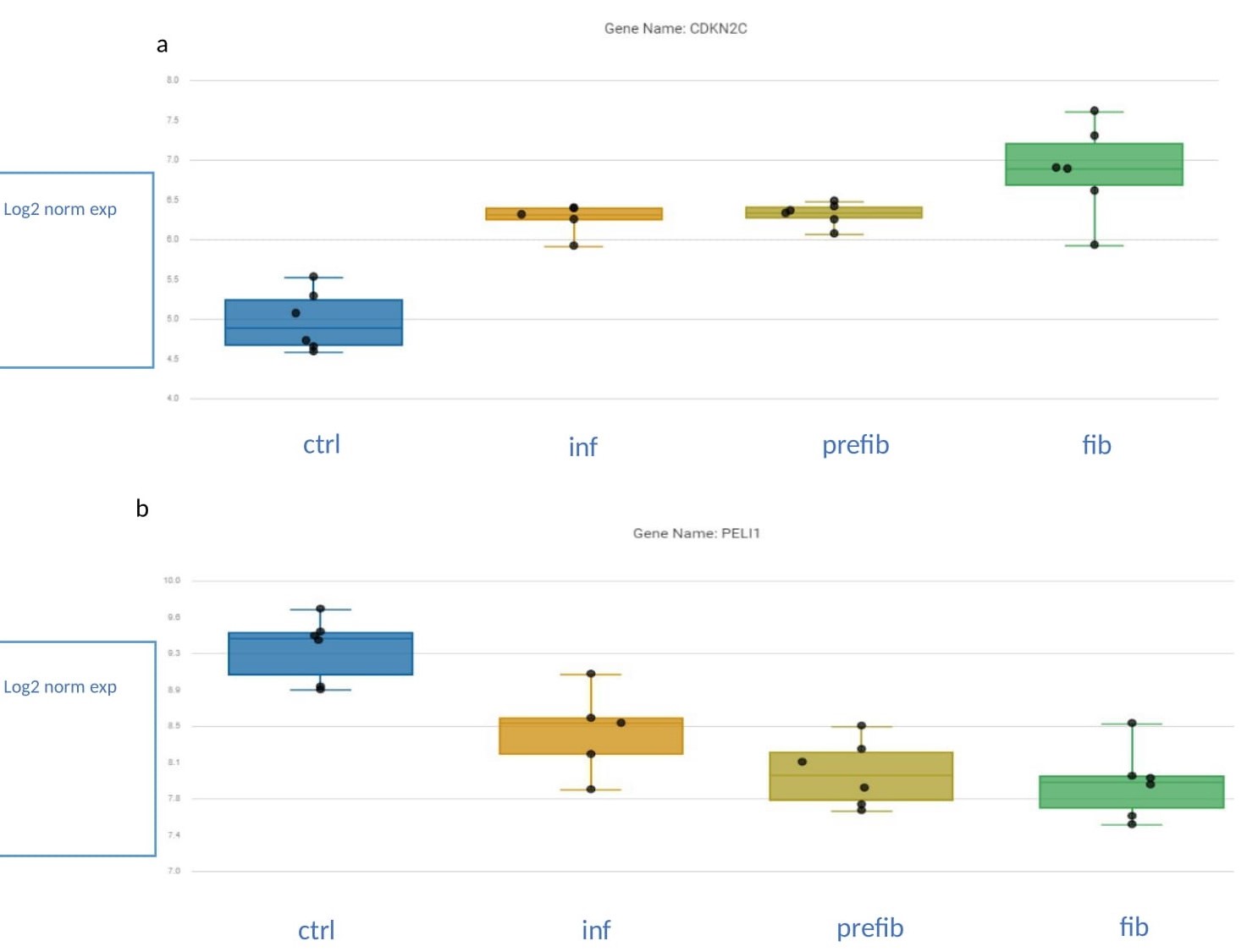
Results: To validate our simulation model, we performed subgroup analysis that showed an incremental increase in pathway scores related to the severity of fibrosis including, among others, ECM and collagen 1 biosynthesis, PDGF signalling, and myofibroblast regulation (Fig.1). Ctrl vs SSc-ILD comparison demonstrated 24 differentially expressed genes, two of which had the most pronounced p-values. Cyclin-Dependent Kinase Inhibitor 2C (CDKN2C) was significantlyoverexpressed (p = 0.00052) in SSc-ILD compared to ctrl, while expression of Pellino E3 ubiquitin-protein ligase 1 (PELI1) showed lower expression significantly (p = 0.0012). Additionally, the expression of CDKN2C and PELI1 showed an incremental increase and decrease in the four groups, respectively (Fig.2). Immunohistochemistry of CDKN2C and PELI1 showed consistent results with the nS analysis with incremental increase and decrease of positive staining in the 4 groups, respectively.
Conclusion: Expression of CDKN2C and PELI1 was associated with more severe stages of SSc-ILD on histologic assessment. PELI1 decrease is probably due to loss of bronchial tissue during the fibrosis processes and thus secondary to the already existing pathology. We report the potential of the cell cycle inhibitor, CDKN2C to predict fibrosis progression. Further research is required to validate these findings and correlate additional cell cycle regulation and/or senescence markers with CDKN2C. determine the potentiality of CDKN2C as a putative progression marker candidate.
To cite this abstract in AMA style:
Al-Adwi Y, Westra J, van Goor H, van Kempen L, Timens W, Gan C, Mulder D. Transcriptomic Analyses of Lung Tissues Reveals Potential Key Genes Associated with Progression of Systemic Sclerosis-Interstitial Lung Disease (SSc-ILD) [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/transcriptomic-analyses-of-lung-tissues-reveals-potential-key-genes-associated-with-progression-of-systemic-sclerosis-interstitial-lung-disease-ssc-ild/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/transcriptomic-analyses-of-lung-tissues-reveals-potential-key-genes-associated-with-progression-of-systemic-sclerosis-interstitial-lung-disease-ssc-ild/