Session Information
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Ankylosing spondylitis (AS) is an autoimmune and auto-inflammatory disease characterized by chronic inflammation of the spine, sacroiliac joint, and occasionally peripheral joints. TNF-α inhibitor (TNFi) are among the primary treatments for AS. However, the precise mechanisms by which they exert their effects are not fully understood. We aimed to find the key genes expressed in the critical immune cell and elucidate their roles in the therapeutic action of TNFi in AS.
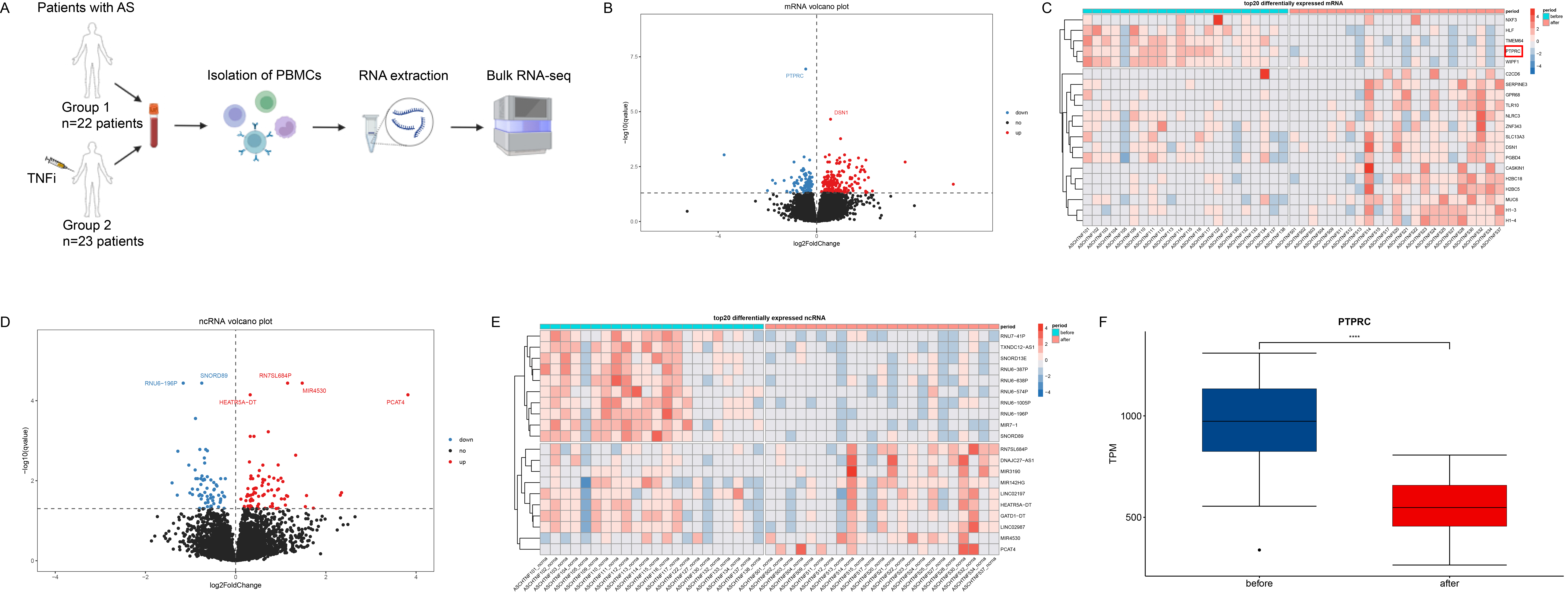
Methods: Bulk RNA-seq on peripheral blood mononuclear cells (PBMCs) was performed on AS patients pre- and post-treatment, including 22 AS patients before treatment with TNFi and 23 AS patients after three months of the treatment (Figure 1A). CIBERSORT was used for deconvolution analysis. Differentially expressed genes (DEGs) analysis was performed by DESeq2. Protein-protein interaction (PPI) analysis was performed by Search Tool for the Retrieval of Interacting Genes/Proteins (STRING). The expression of key genes in immune cells was searched on the Human Protein Atlas (HPA) database. Based on Spearman correlation analysis, the mRNA-non-coding RNA(ncRNA) co-expression network was built in Cytoscape. Gene set enrichment analysis (GSEA) was applied for pathway analysis.
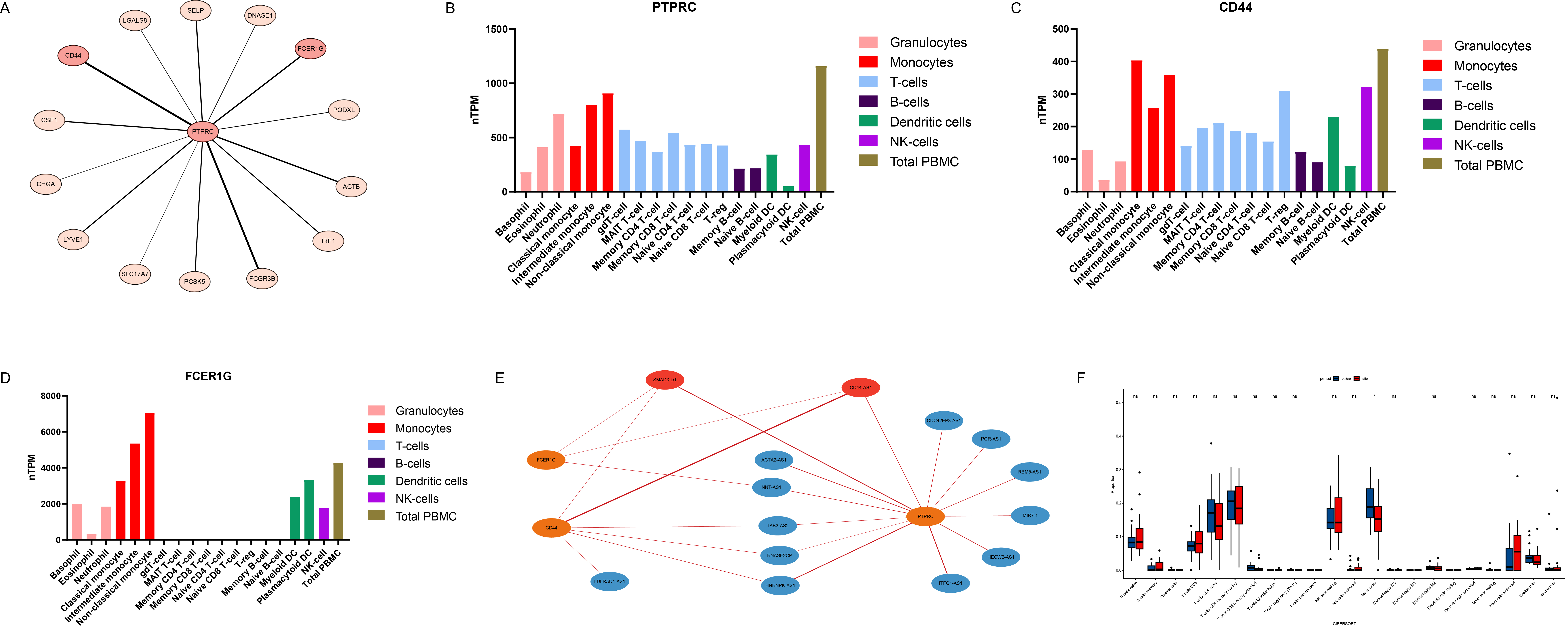
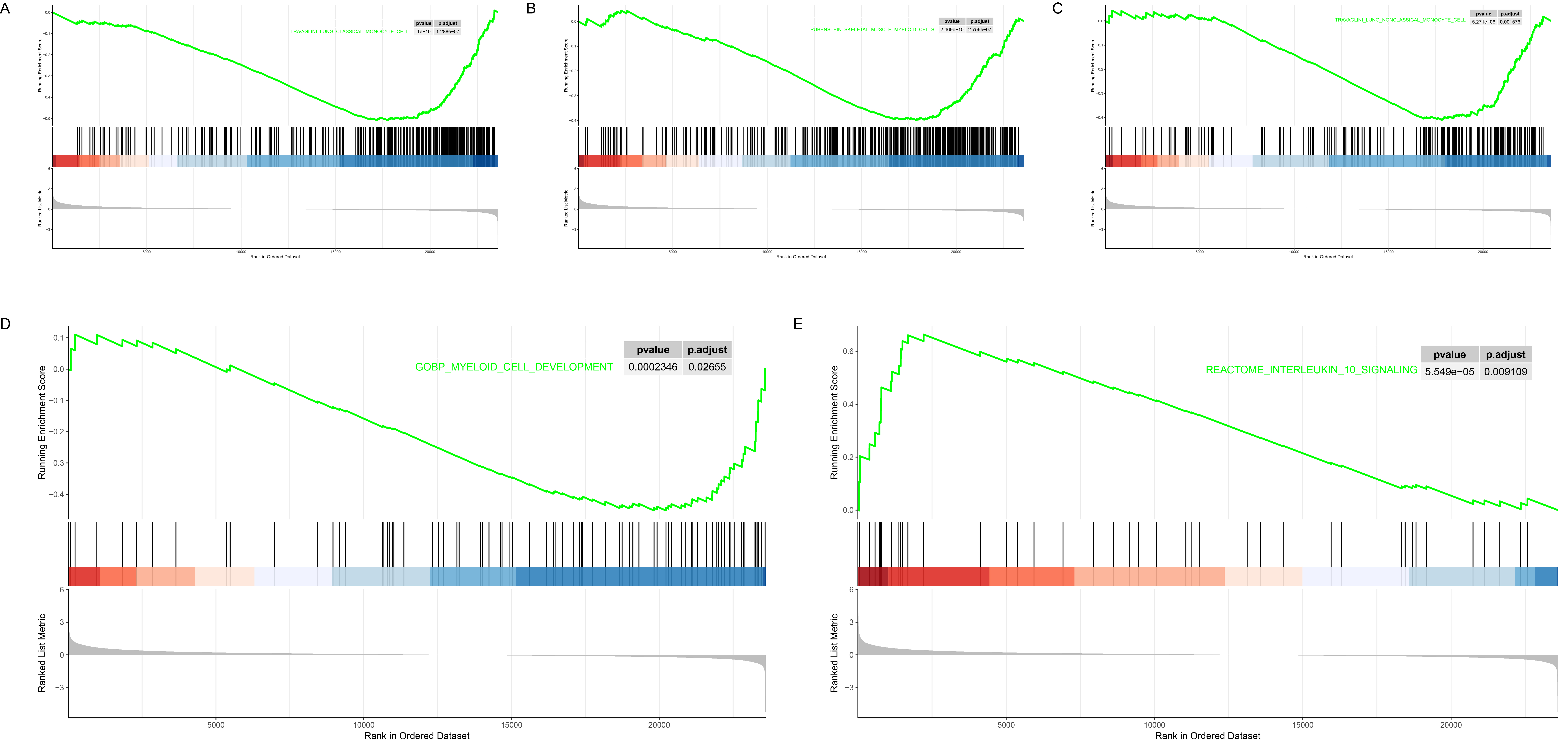
Results: We identified 466 differentially expressed genes (DEGs), consisting of 320 mRNA and 146 ncRNA (Figures 1B-E). Among these, PTPRC emerged as the most significantly expressed gene (Figure 1F). Protein-protein interaction (PPI) analysis revealed that PTPRC strongly interacted with CD44 and FCER1G (Figure 2A), both of which were highly expressed in monocytes (Figures 2B-D). Our mRNA-ncRNA co-expression network analysis suggested that certain long non-coding RNAs (lncRNAs), such as SMAD3-DT and CD44-AS1, might regulate the expression of PTPRC, CD44, and FCER1G (Figure 2E). Deconvolution analysis also showed that a significant down-regulation in the proportion of monocytes following TNF inhibitor (TNFi) treatment (Figure 2F). Moreover, gene set enrichment analysis (GSEA) indicated pathways related to innate immunity and inflammation, including myeloid cell differentiation, were significantly down-regulated post-treatment (Figures 3A-D). On the contrary, anti-inflammatory pathways such as “interleukin-10 signaling” were significantly up-regulated after the treatment (Figure 3E).
Conclusion: Our study reveals the important role of monocyte-related innate immunity in treating AS with TNFi. Genes like PTPRC, CD44 and FCERlG are highly expressed in monocytes and may play key roles in drug response via the regulatory lncRNA such as SMAD3-DT and CD44-AS1. These molecules are expected to decipher the mechanisms of TNFi therapy and predict its efficacy.
To cite this abstract in AMA style:
Tang Y, Xie J, Zhuo D, ZHU Q, Wang J, Liu J. Transcriptome Analysis Characterizes the Role of Monocytes in Ankylosing Spondylitis with TNF-blocker Treatment [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/transcriptome-analysis-characterizes-the-role-of-monocytes-in-ankylosing-spondylitis-with-tnf-blocker-treatment/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/transcriptome-analysis-characterizes-the-role-of-monocytes-in-ankylosing-spondylitis-with-tnf-blocker-treatment/