Session Information
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Understanding pathogenic processes in the diseased tissue in rheumatoid arthritis (RA) is a critical step to defining disease pathogenesis and identifying novel therapeutic targets. However, the use of enzymatically dispersed cells can lead to changes in the gene transcription and bias due to cell death. Therefore, we established laser capture microscopy followed by RNAseq (LCM-RNAseq) to study the regional transcriptome.
Methods: Synovial tissue was captured by LCM after H&E staining from lining, sublining, and blood vessels from 3 RA and 3 osteoarthritis (OA) patients. RNA was extracted and subjected to RNAseq. Principal component analysis (PCA) by disease state was performed on the top 500 most variable genes among the sample set. Analysis of differentially expressed genes (DEGs) between disease state was performed using the Bioconductor DESeq2 package. DEGs had adjusted p-values of < 0.05 and log2 fold change >1 or < -1. Pathway analysis on the unique DEGs for each substructure was performed using the Canonical Pathways function in Ingenuity Pathway Analysis (IPA) platform. The significance of the pathways was calculated using a right-tailed Fisher’s exact test and adjusted using the Benjamini-Hochberg method.
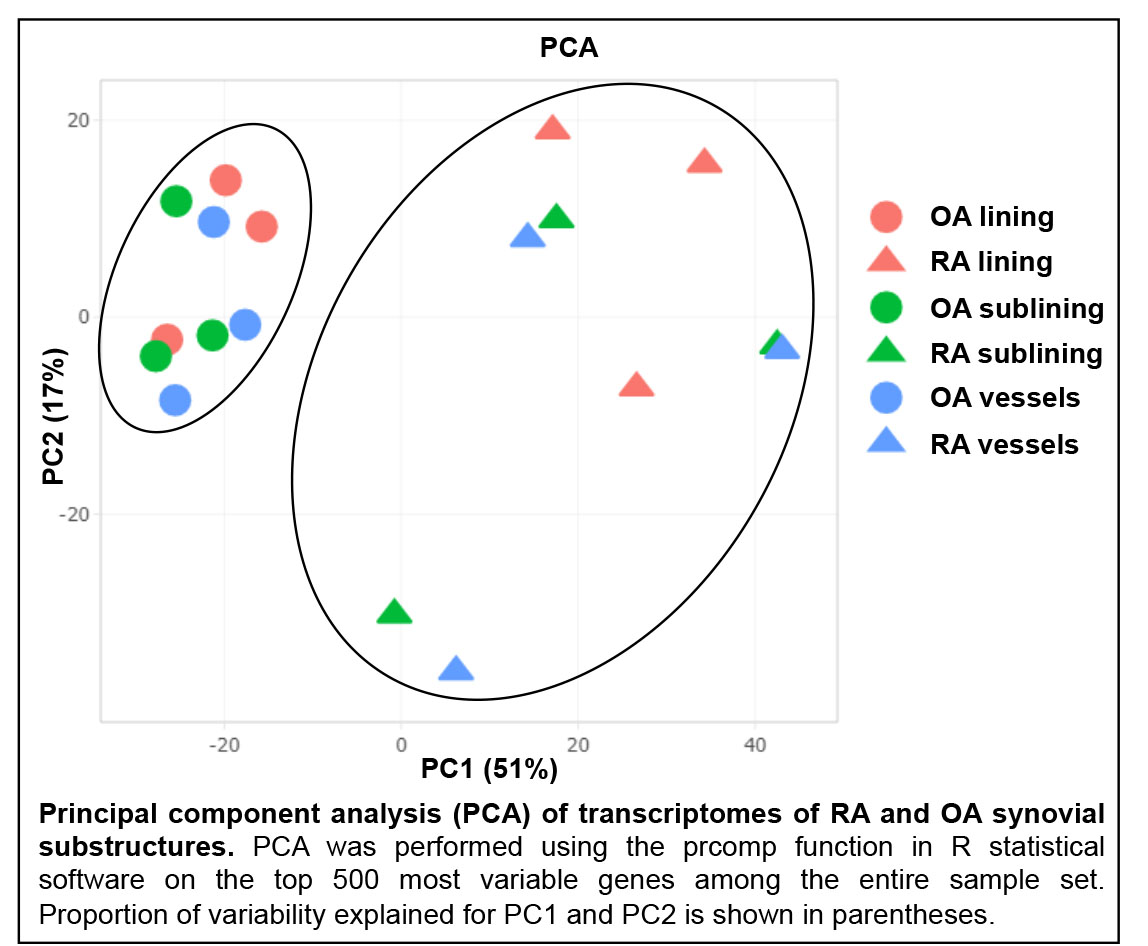
Results: PCA revealed striking separation between RA and OA transcriptomes (see Figure), indicating that the transcriptomes could be readily distinguished. Pairwise comparisons of synovial lining, sublining and blood vessels between RA and OA revealed substantial differences in transcriptional patterns between each region. Pathway analysis of DEGs unique to each substructure revealed that each region displayed distinct phenotypic abnormalities in RA. For example, RA synovial lining was marked by anomalies of innate immunity, including Role of Pattern Recognition Receptors in Recognition of Bacteria and Virus (p< 0.001) and LPS/IL-1 Mediated Inhibition of RXR Function (p< 0.01) pathways. Intriguingly, RA sublining abnormalities were primarily in metabolic pathways involving fatty acid regulation, such as Fatty Acid Activation (p< 0.01) and Mitochondrial L-carnitine Shuttle Pathway (p< 0.01). RA synovial vessels displayed evidence of alterations in pathways indicative of dysregulated angiogenesis, including Melatonin Signaling (p< 0.0001) and Apelin Cardiomyocyte Signaling (p< 0.001).
Conclusion: Through use of LCM, we established a method for RNAseq-quality extraction of RNA and showed large differences between RA and OA synovial substructures that are difficult to detect using single cell analysis. This technology can supplement disaggregation techniques to provide key information on in situ gene expression profiles and identify disease and substructure-specific targets for RA.
To cite this abstract in AMA style:
Van Espen B, Wilson A, Seumois G, Perumal N, Benschop R, Firestein G, Bottini N, Stanford S. Topological Laser Capture Microscopy (LCM)-RNAseq to Map the Rheumatoid Arthritis (RA) Synovial Transcriptome [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/topological-laser-capture-microscopy-lcm-rnaseq-to-map-the-rheumatoid-arthritis-ra-synovial-transcriptome/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/topological-laser-capture-microscopy-lcm-rnaseq-to-map-the-rheumatoid-arthritis-ra-synovial-transcriptome/