Session Information
Session Type: ACR Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose:
A
previously described transcriptional signature present in circulating CD4+ T
cells of early rheumatoid arthritis (RA) patients implicated STAT3 signalling
as an early pathophysiological event1. We sought to validate this
finding through independent replication and microarray meta-analysis.
Methods:
Microarray
technology was used to measure gene expression in highly purified peripheral
blood CD4+ T cells from treatment-naïve RA patients and disease controls recruited
from an early arthritis clinic. Analysis focussed on 12 transcripts identified
during the previous study, and concurrent STAT3 pathway activation was determined
in the same cells by flow cytometry. A meta-analysis analysis of previous and
current gene expression findings employed multivariate and receiver operator
characteristic curve analysis.
Results:
In
the independent cohort of 161 patients, normalised expression of 11 of the 12 original
signature genes differed significantly between RA patients and controls. Differential
regulation was most pronounced for the STAT3 target genes PIM1, BCL3 and SOCS3
(>1.3-fold difference; p<0.004), each of whose expression correlated strongly
with paired intracellular phospho-STAT3, and these 3 genes’ expression
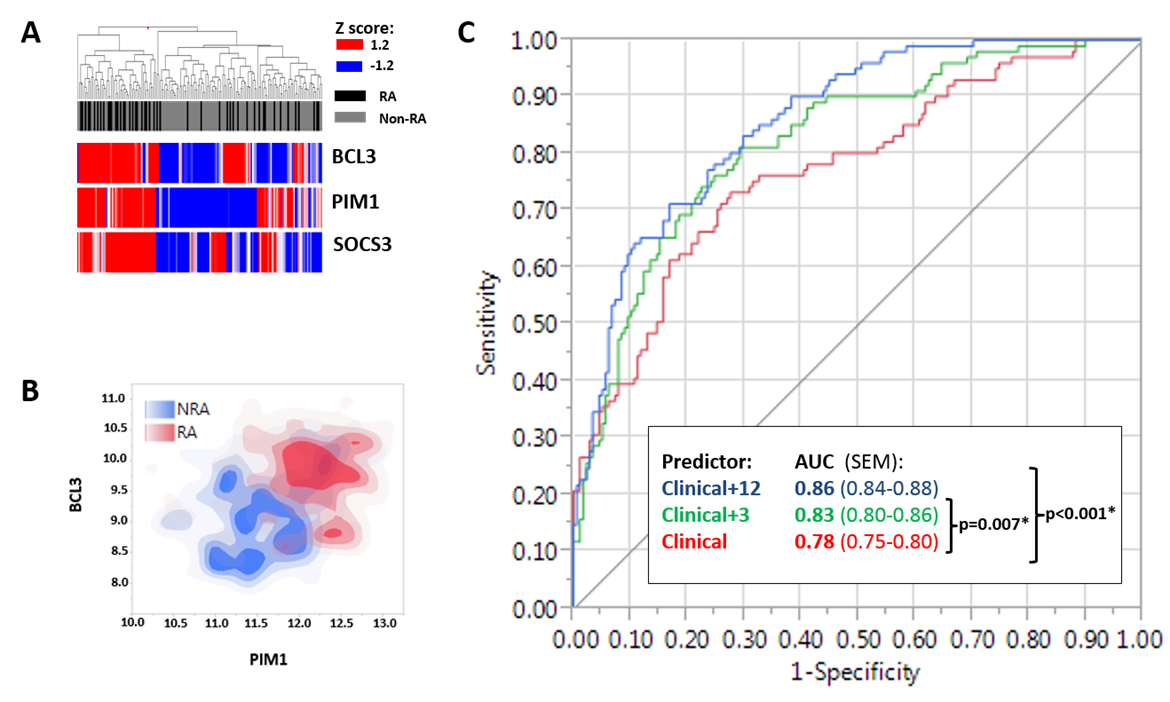
accounted for the majority of the signature’s ability to segregate diagnoses by
hierarchical clustering (Figure 1A). The ability of the same 3 genes, or pairs
thereof, to segregate RA patients from disease controls was confirmed in the meta-analysis
of 279 patients (E.g. Figure 1B). Moreover, both the 12 gene and the 3 gene
signatures added independent discriminatory value to a consideration of
clinical parameters including age, swollen/tender joint counts and acute phase
response with respect to diagnosis (Figure 1C).
Conclusion:
The
STAT3 mediated dysregulation of BCL3, SOCS3 and PIM1 in circulating CD4+ T
cells is confirmed as an independent early event in RA pathogenesis. The
mechanistic and functional implications of this observation are being
investigated.
Figure
1. A. Hierarchical
clustering based on 3 genes’ expression in T cells discriminates early RA
patients from disease controls. B Non-parametric density plot depicting
utility of BCL3 and PIM1 alone in discriminating RA patients. C. Adding
either a 3 gene or 12 gene CD4+ T cell “signature” to a predictive model based
on clinical parameters alone significantly enhanced discrimination of RA from
non-RA.
1.
Pratt AG et al. 2012;
Annals of the Rheumatic Diseases
To cite this abstract in AMA style:
Pratt AG, Anderson AE, Lendrem DW, Skelton A, Massey J, Nair N, Diboll J, Hargreaves B, Brown PM, Barton A, Isaacs JD. STAT3-Regulated Gene Expression in Circulating CD4+ T Cells Discriminates RA Patients Independently of Clinical Parameters in Early Arthritis: A Validation Study [abstract]. Arthritis Rheumatol. 2015; 67 (suppl 10). https://acrabstracts.org/abstract/stat3-regulated-gene-expression-in-circulating-cd4-t-cells-discriminates-ra-patients-independently-of-clinical-parameters-in-early-arthritis-a-validation-study/. Accessed .« Back to 2015 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/stat3-regulated-gene-expression-in-circulating-cd4-t-cells-discriminates-ra-patients-independently-of-clinical-parameters-in-early-arthritis-a-validation-study/