Session Information
Date: Sunday, October 26, 2025
Title: (0067–0097) Rheumatoid Arthritis – Etiology and Pathogenesis Poster
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: RA synovium displays cellular heterogeneity, with gene expression driving disease pathogenesis. Unbiased cell-specific transcriptomes in RA synovium have previously relied primarily on disaggregated tissues to understand these pathogenic processes, however, this approach can cause cell loss and gene induction. Therefore, we interrogated RA and OA synovial tissue using spatial transcriptomics to investigate spatial and cell-type differences, focusing on lining and sublining regions segmented into fibroblasts and macrophages.
Methods: Fresh frozen synovial tissues from 7 RA and 8 OA patients were prepared for the NanoString GeoMX DSP Whole Transcriptome Assay. Lining and sublining regions were segmented to fibroblasts (vimentin+ CD68-) and macrophages (vimentin− CD68+) (Fig.1). NovaSeq 6000 was used for sequencing. 121 segments passed QC, with 10,363 genes detected and Q3-normalized. A logistic regression (LR) classifier was trained on principal components to find the top 500 genes separating RA from OA. Differentially expressed genes (DEG) were analyzed by linear mixed models (p-value< 0.05 and log2(FC) >0.5), and Reactome pathway analysis (FDR< 0.02).
Results: LR models separation RA from OA found discriminative genes enriched in inflammation and tissue remodeling pathways (e.g., TNF/IL17/IFN signaling, ECM). DEG analysis showed distinct RA vs OA gene signatures across regions and cell types (Fig.2). RA lining fibroblasts had 700 DEG compared with OA lining, with enrichment for 18 pathways that were focused on inflammation and matrix regulation (see Fig.3). RA sublining fibroblasts showed 834 DEG with OA sublining, with enrichment for 29 pathways. Although some inflammatory pathways were identified, sublining fibroblasts also included multiple pathways related to regulation of adaptive immunity (see Fig.3). Spatial comparisons between lining and sublining in RA showed that lining fibroblasts displayed 1176 DEG with 37 enriched pathways, including RAC1 GTPase cycle, collagen biosynthesis/modifying enzymes, and post-translational protein phosphorilation. Therefore, RA lining fibroblasts display a highly activated phenotype compared with sublining cells and help define the aggressive biology of fibroblast-like synoviocytes which are a hallmark in RA. RA lining macrophages showed 628 DEG compared to OA lining, enriched for 60 pathways (e.g., ECM organization, IFN gamma signaling, translation). Surprisingly, even though RA sublining macrophages had 672 DEGs compared to OA, they were only enriched in 2 pathways, suggesting that the differences are random and that sublining macrophages in RA might not drive pathology.
Conclusion: Spatial transcriptomic analysis reveals distinct region- and cell-type-specific transcriptional profiles distinguishing RA from OA synovium. Regional analysis identifies niche-specific fibroblast functions related to inflammation in the lining and adaptive immunity in the sublining were identified, which could help identify subsets as therapeutic targets for patient-specific mechanisms of disease.
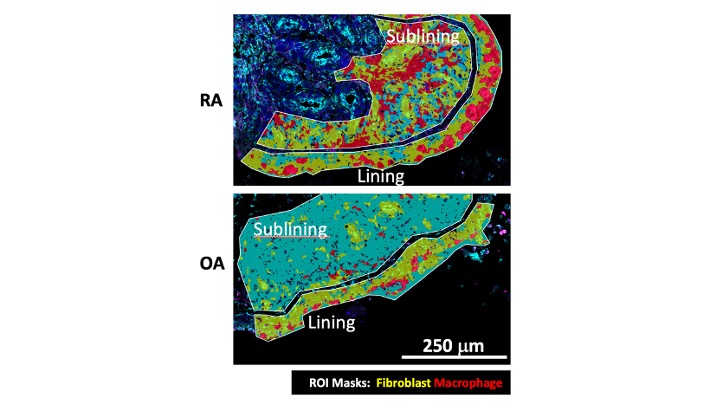 Figure 1. Spatial transcriptomics analysis using NanoString GeoMX DSP Whole Transcriptome Assay. Panel shows examples of RA and OA synovial tissues stained and visualized for fibroblasts (vimentin; yellow-green) and macrophages (CD68; red). The lining and sublining regions were identified as regions of interest (ROI) to quantify gene expression normalized to cell lineage.
Figure 1. Spatial transcriptomics analysis using NanoString GeoMX DSP Whole Transcriptome Assay. Panel shows examples of RA and OA synovial tissues stained and visualized for fibroblasts (vimentin; yellow-green) and macrophages (CD68; red). The lining and sublining regions were identified as regions of interest (ROI) to quantify gene expression normalized to cell lineage.
.jpg) Figure 2. Volcano plots of differentially expressed genes between OA and RA groups for macrophages and fibroblasts in lining and sublining regions. DEGs were identified using linear mixed models (LMM) with a significance threshold of p-value < 0.05 and log2(FC) > 0.5. DEGs in blue are significant genes enriched in RA or OA.
Figure 2. Volcano plots of differentially expressed genes between OA and RA groups for macrophages and fibroblasts in lining and sublining regions. DEGs were identified using linear mixed models (LMM) with a significance threshold of p-value < 0.05 and log2(FC) > 0.5. DEGs in blue are significant genes enriched in RA or OA.
.jpg) Table 1. Different enriched pathways in lining and sublining fibroblasts from synovial tissue of patients with RA versus OA. Pathways highlighted in bold represent those related to the adaptive immune system. The Reactome pathway analysis was performed using differentially expressed genes in the RA versus OA in lining and sublining fibroblasts (p < 0.05). The table displays only pathways that were significantly enriched exclusively in either lining or sublining fibroblasts.
Table 1. Different enriched pathways in lining and sublining fibroblasts from synovial tissue of patients with RA versus OA. Pathways highlighted in bold represent those related to the adaptive immune system. The Reactome pathway analysis was performed using differentially expressed genes in the RA versus OA in lining and sublining fibroblasts (p < 0.05). The table displays only pathways that were significantly enriched exclusively in either lining or sublining fibroblasts.
To cite this abstract in AMA style:
Machado C, Yao M, Boyle D, Benschop R, Parker J, Wang W, Firestein G. Spatial transcriptomics in rheumatoid arthritis (RA) synovium reveals distinct region-specific fibroblast functions [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/spatial-transcriptomics-in-rheumatoid-arthritis-ra-synovium-reveals-distinct-region-specific-fibroblast-functions/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/spatial-transcriptomics-in-rheumatoid-arthritis-ra-synovium-reveals-distinct-region-specific-fibroblast-functions/
