Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Uncharacterized microbial enzymes in metagenomics are difficult to annotate, especially in fibrosis-prone conditions like IgG4-related disease (IgG4-RD) and Crohn’s disease (CD), where microbial carbohydrate metabolism may influence disease. Traditional sequence homology-based tools often miss enzymes with low sequence similarity. Protein language models (pLMs) provide a new AI-driven approach. We present CAZyLingua, the first pLM-based deep learning tool for CAZyme annotation. Applied to IgG4-RD and CD metagenomes, CAZyLingua uncovered hundreds of previously unannotated CAZymes-especially carbohydrate esterases (CEs)-broadening our understanding of the microbial enzymatic landscape in fibrotic diseases.
Methods: CAZyLingua was trained on a non-redundant CAZy database clustered at 60% sequence identity, with sequence embeddings generated using ProtT5 pLM. The pipeline used a quadratic discriminant analysis (QDA) classifier for CAZyme detection, followed by a four-layer neural network for family and subfamily classification, employing weighted cross-entropy loss to address class imbalance. Hyperparameters were optimized with RayTune across 100 epochs and 20 parallel models. Performance was benchmarked against dbCAN2 using three gold-standard bacterial genomes. CAZyLingua was then applied to metagenomic gene catalogs from CD and IgG4-RD, with linear modeling to identify differentially abundant CAZymes. Functional validation of a predicted CE17 from the CD cohort was performed via recombinant protein expression and MALDI-ToF mass spectrometry.
Results: CAZyLingua outperformed dbCAN2 in precision, recall, and F1 score for CAZyme identification, with up to 10% improvement in F1 for certain strains. In disease-associated metagenomes, CAZyLingua identified hundreds of CAZymes not detected by dbCAN2, with notable enrichment of CE families-especially CE1, CE3, CE4, CE12, and CE17. In IgG4-RD, CAZyLingua predicted 437 additional CAZymes, 34% of which were CEs, a class underrepresented in reference databases. In CD, CAZyLingua identified a subset of CAZymes more abundant in disease, including CE17. Functional assays confirmed that CE17 catalyzed deacetylation of acetylated mannooligosaccharides, validating the model’s annotation and the biological relevance of these newly identified enzymes.
Conclusion: CAZyLingua leverages pLMs to expand CAZyme annotation in fibrotic disease metagenomes, uncovering rare and structurally divergent enzymes overlooked by homology-based methods. The tool’s ability to predict and validate novel enzymatic activities, such as CE17 in CD, demonstrates its value for elucidating the microbiome’s role in fibrosis and inflammation. In IgG4-RD, CAZyLingua identified hundreds of additional CAZymes-particularly CEs-missed by standard tools, revealing an expanded repertoire of microbial enzymes potentially relevant to disease pathogenesis. These findings highlight CAZyLingua’s utility for uncovering hidden microbial functions in both CD and IgG4-RD, supporting its broader application for comprehensive protein function discovery in fibrosis-prone disease states.
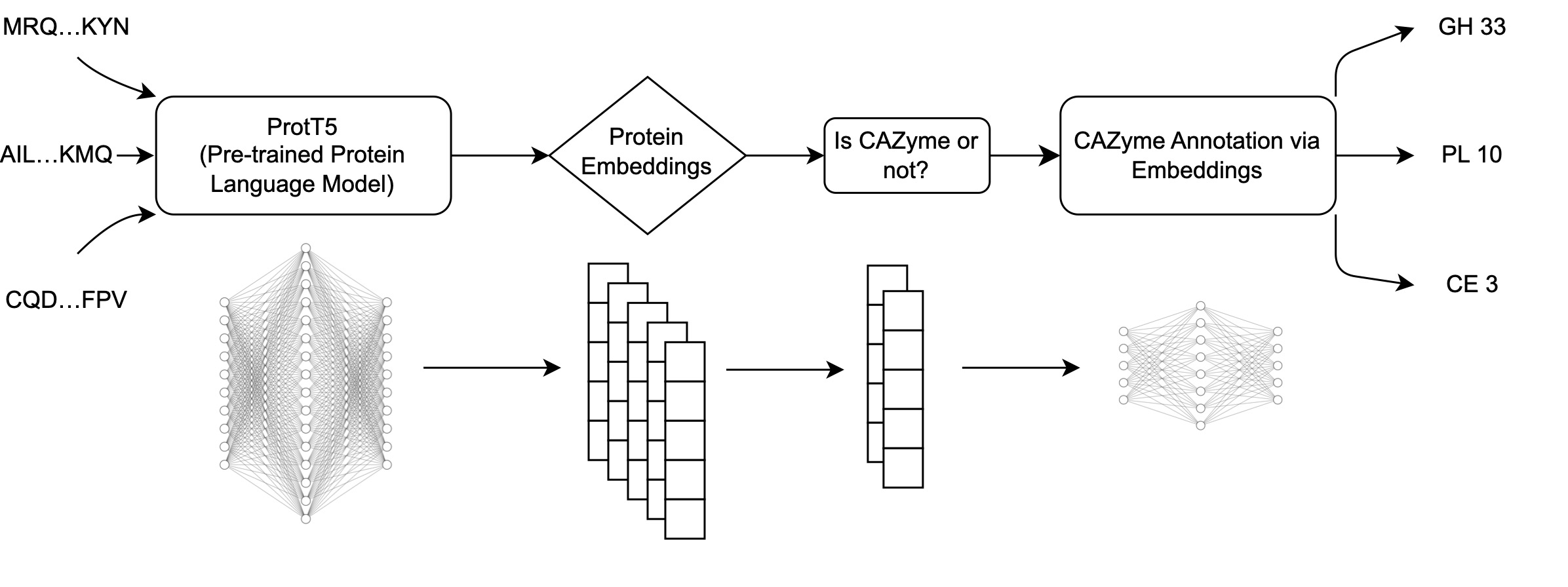 The workflow of CAZyLingua starts with raw embeddings from ProtT5 followed by the use of those embeddings as input through two classifiers to distinguish 1) whether the embedding was a CAZyme and if so, 2) to which CAZyme family it belongs to.
The workflow of CAZyLingua starts with raw embeddings from ProtT5 followed by the use of those embeddings as input through two classifiers to distinguish 1) whether the embedding was a CAZyme and if so, 2) to which CAZyme family it belongs to.
.jpg) a) Genes enriched and depleted in the gene catalogs of patients with IgG4-RD selected on the fringe of the volcano plot (see Methods for labeling criteria). b) Predicted CEs in the enriched IgG4-RD gene set, stratified to analyze only the genes CAZyLingua predicted. c) The proportion of dbCAN2-predicted CAZymes also predicted by CAZyLingua as the decision function between CAZyme/non-CAZyme of the QDA classifier in CAZyLingua was varied. The Venn diagram shows the numbers of CAZymes predicted by CAZyLingua, dbCAN2, and both on our current model benchmarks of the QDA. d) Genes enriched and depleted in the gene catalogs of patients with CD selected on the fringe of the volcano plot (see Methods for labeling criteria). CE17 is highlighted in the circle.
a) Genes enriched and depleted in the gene catalogs of patients with IgG4-RD selected on the fringe of the volcano plot (see Methods for labeling criteria). b) Predicted CEs in the enriched IgG4-RD gene set, stratified to analyze only the genes CAZyLingua predicted. c) The proportion of dbCAN2-predicted CAZymes also predicted by CAZyLingua as the decision function between CAZyme/non-CAZyme of the QDA classifier in CAZyLingua was varied. The Venn diagram shows the numbers of CAZymes predicted by CAZyLingua, dbCAN2, and both on our current model benchmarks of the QDA. d) Genes enriched and depleted in the gene catalogs of patients with CD selected on the fringe of the volcano plot (see Methods for labeling criteria). CE17 is highlighted in the circle.
.jpg) e) The enriched genes in CD predicted by CAZyLingua only were prioritized based on a combination of the log fold change and the probability of the CAZyme annotation from CAZyLingua. The plot is ordered from the highest fold change and CAZyLingua prediction probability (red) to the lowest fold change and prediction probability (blue). CE17 is highlighted in bold. f) Functional characterization of CE17 using MALDI-ToF mass spectrometry. Peaks are labeled by degree of polymerization (DP) and number of acetyl (Ac) groups. The annotated m/z values indicate sodium adducts. Intensity is shown in arbitrary units (a.u.). Both the KTCE17 enzyme (middle) and a previously validated CE17, FpCE17 (bottom, (60)) showed the same activity on a RiGH26-pretreated β-mannan substrate, with disappearance of peaks signifying double and triple acetylated oligosaccharides, and decrease in the intensities of peaks signifying mono-acetylated oligosaccharides (containing 3-O-acetylations) and accumulation of deacetylated oligosaccharides.
e) The enriched genes in CD predicted by CAZyLingua only were prioritized based on a combination of the log fold change and the probability of the CAZyme annotation from CAZyLingua. The plot is ordered from the highest fold change and CAZyLingua prediction probability (red) to the lowest fold change and prediction probability (blue). CE17 is highlighted in bold. f) Functional characterization of CE17 using MALDI-ToF mass spectrometry. Peaks are labeled by degree of polymerization (DP) and number of acetyl (Ac) groups. The annotated m/z values indicate sodium adducts. Intensity is shown in arbitrary units (a.u.). Both the KTCE17 enzyme (middle) and a previously validated CE17, FpCE17 (bottom, (60)) showed the same activity on a RiGH26-pretreated β-mannan substrate, with disappearance of peaks signifying double and triple acetylated oligosaccharides, and decrease in the intensities of peaks signifying mono-acetylated oligosaccharides (containing 3-O-acetylations) and accumulation of deacetylated oligosaccharides.
To cite this abstract in AMA style:
Thurimella K, Mohamed A, Li C, Vatanen T, Graham D, Owens R, Leanti La Rosa S, Plichta D, Bacallado S, Xavier R. Protein Language Model-Guided Homology Identifies Microbial Enzymes Linked to Fibrosis-Prone IgG4-RD and Crohn’s Disease [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/protein-language-model-guided-homology-identifies-microbial-enzymes-linked-to-fibrosis-prone-igg4-rd-and-crohns-disease/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/protein-language-model-guided-homology-identifies-microbial-enzymes-linked-to-fibrosis-prone-igg4-rd-and-crohns-disease/
