Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Pulmonary hypertension (PH) portends poor outcomes in Systemic Sclerosis (SSc). Recent efforts for early identification and intervention in SSc patients with pulmonary arterial hypertension (PAH) with algorithms like DETECT have drastically improved outcomes in this population. However, such algorithms have failed in other WHO (World Health Organization) PH subgroups. This study aims to evaluate the effectiveness of the DETECT algorithm and Cluster analysis in predicting Group 2 (PH due to left heart disease) and Group 3 (PH due to chronic lung disease).
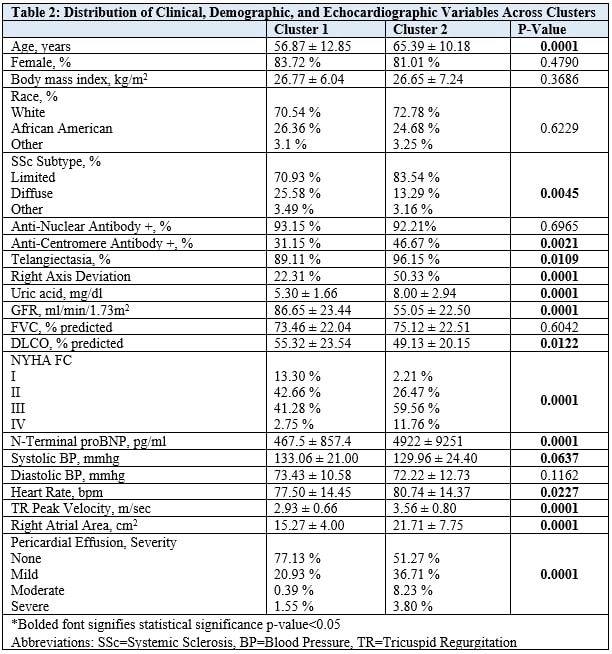
Methods: We included a well-characterized cohort of SSc patients from our center with clinical, laboratory, and echocardiographic data near the right heart catheterization date. Scores were generated for each step of the DETECT algorithm when applicable. Then, k-means cluster analysis was conducted using clinical, laboratory, and echocardiographic variables. Table 2. Cluster characteristics were compared through the Kruskal-Wallis test. Clustering was decided according to the elbow plot. ROC analysis across all three PH groups was then conducted for step 1 of the DETECT algorithm, using an empirically optimal cutoff. For the cluster model, ROC analysis considered inclusion in cluster 2 as a positive screening test, while inclusion in cluster 1 was considered negative.
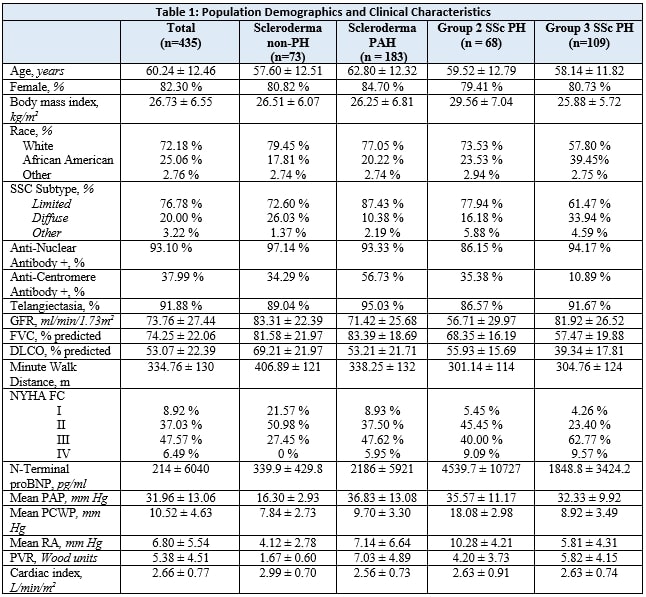
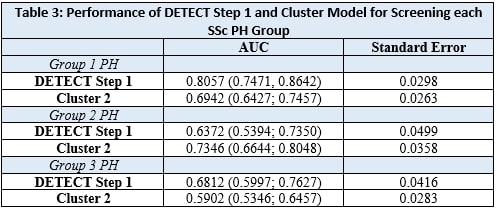
Results: Our cohort consisted of 435 SSc patients, 183 with PAH, 68 with Group 2 PH, 109 with Group 3 PH, and 73 without PH: mean age 60 ± 12 years, 82% female, 72% White, 76.8% limited SSc. Table 1. Across PH Groups, AUC ranged from 0.68 – 0.81, with SSc- PAH having the greatest AUC. Table 3. The DETECT algorithm’s performance in Group 2 SSc PH (AUC = 0.64) and Group 3 SSc PH (AUC = 0.68) was considerably less reliable than in SSc PAH (AUC = 0.81). Variables chosen for cluster analysis are all listed in Table 2. The clustering model did not perform adequately in predicting PAH (AUC = 0.69) or Group 3 PH (AUC = 0.59); however, it performed better than DETECT in screening for Group 2 PH (AUC = 0.73). Table 3.
Conclusion: The DETECT algorithm performs poorly in screening for SSc Group 2 and Group 3 PH. Patient clustering provides improved performance in predicting Group 2 PH but was not effective in predicting PAH or Group 3 PH.
To cite this abstract in AMA style:
Osgueritchian R, Mombeini H, Jani V, Woods A, Hsu S, Lammi M, Hassoun P, Wigley F, Hummers L, Mathai S, Shah A, Mukherjee M. Performance of the DETECT Algorithm and Cluster Analysis in Screening for Systemic Sclerosis Pulmonary Hypertension Groups [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/performance-of-the-detect-algorithm-and-cluster-analysis-in-screening-for-systemic-sclerosis-pulmonary-hypertension-groups/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/performance-of-the-detect-algorithm-and-cluster-analysis-in-screening-for-systemic-sclerosis-pulmonary-hypertension-groups/