Session Information
Session Type: Poster Session A
Session Time: 1:00PM-3:00PM
Background/Purpose: Synovial pathotyping is a critical step in understanding the etiology of inflammatory arthritis as different pathotypes have differential response to therapy. Determining these pathotypes relies on immunohistochemistry of lymphoid, myeloid and stromal cells with expert grading of histologic sections, which can be time consuming and costly. Here, we developed an automated segmentation and cell type classification pipeline to efficiently phenotype a murine inflammatory arthritis model; and demonstrate the feasibility of this analysis in human synovial biopsies.
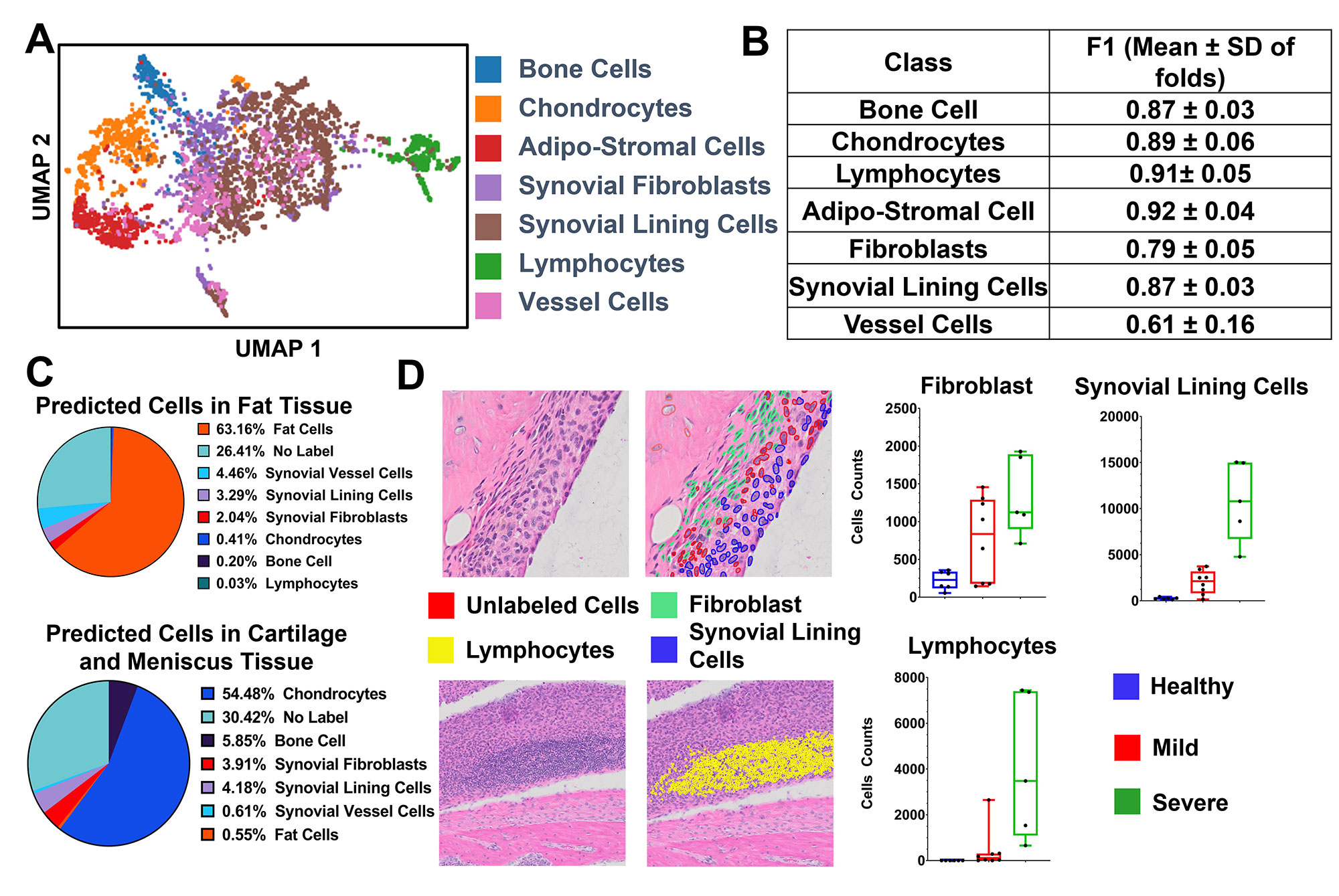
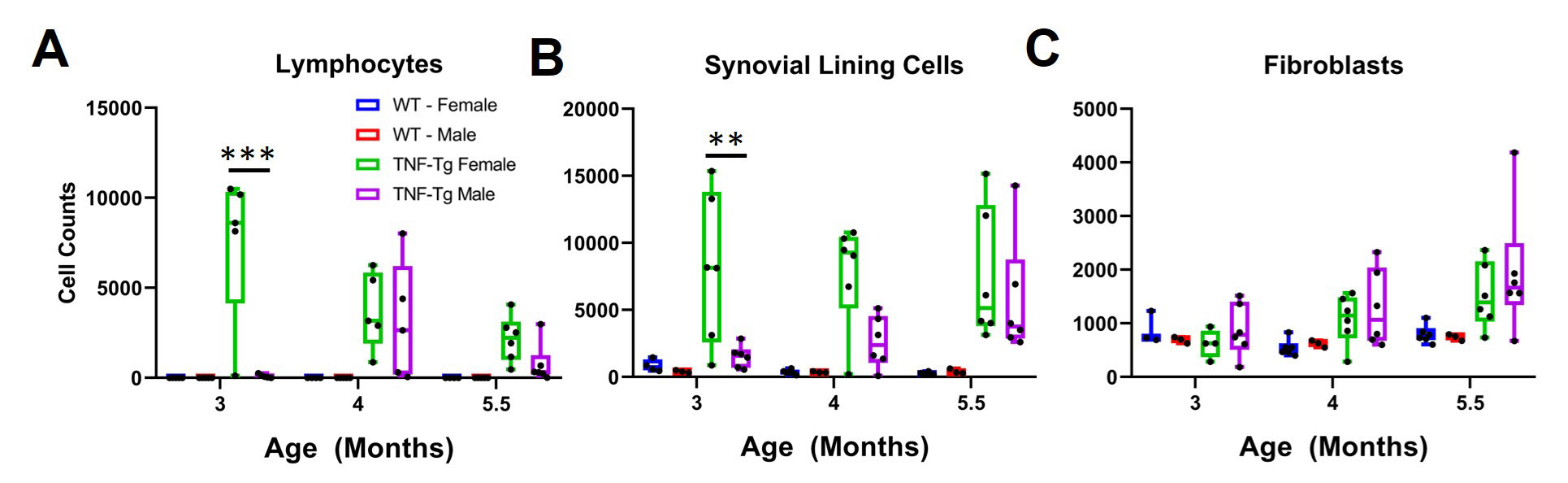
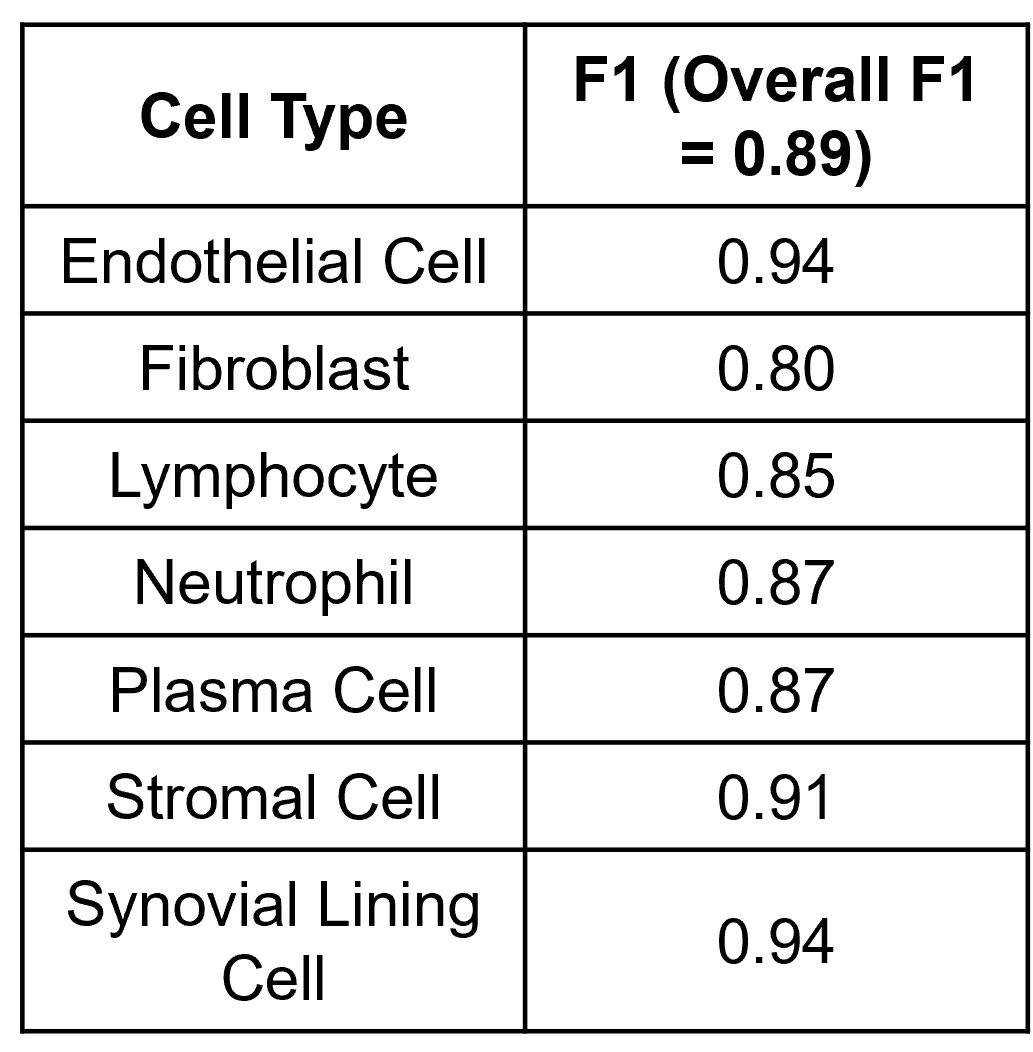
Methods: The synovial tissue, cartilage, bone, and fatty tissue from H&E sections of healthy (n=6), mild (n=8) and severely diseased (n=5) TNF transgenic mice that develop arthritis (Training and Testing Slide) was segmented using a knee tissue segmentation model previously developed in our lab1. We then annotated bone cells (n=312), vessel cells (n=378), adipo-stromal cells (n=506), fibroblasts (n=749), chondrocytes (n=625), lymphocytes (n=467), and all other synovial lining cells (n=1675). Nuclei were segmented and cellular image features were extracted in ImageJ/Python. A stratified 5-fold cross validation strategy was used to train a Gradient Boosted Decision Tree to classify cell types and F1 scores were calculated. Another set of 68 TNF-Tg slides2 (External Validation) describing the model’s sexual dimorphism were subjected to the tissue segmentation, nuclear segmentation, and cell feature extraction and cell types from the synovial tissue were inferred to validate the ability to phenotype the synovium. Also, five human synovial biopsies H&E sections from the Accelerating Medicine Partnership – Rheumatoid Arthritis consortium were annotated for 7 cell types: endothelial cell (n=117), fibroblasts (n=61), lymphocytes (n=71), neutrophils (n=93), plasma cells (n=77), stromal cells (n=57) and synovial lining cells (n=26). The same image analysis and machine learning pipeline was performed.
Results: UMAP demonstrated good separation of TNF-Tg cells and our classification model performed well (Fig 1A, B). We inferred the cell types of the remaining cells on the training and testing slides and demonstrated the tissue specificity of adipo-stromal cells and chondrocytes as the major cell type within their respective tissue (Fig1 C). When stratified by disease severity, fibroblasts, lining cells and lymphocytes followed the expected distribution (Fig 1D). In the external validation set, we also observed the expected increase in synovial lymphocytes and stromal cells in TNF-Tg female mice compared to males at earlier timepoints, but interestingly not in the fibroblast compartment, which suggested a differential etiology (Fig 2A-C). Our pipeline is also able to confidently classify 7 cell types in human RA synovial biopsies, suggesting that we might be able to incorporate our process into other synovial pathotyping pipelines (Fig 3).
Conclusion: We developed a computational pathology cell classification pipeline that confidently identifies cell types in murine and human inflammatory arthritis H&E stained tissue sections. This will allow for efficient pathotyping of diseased tissues to elucidate varying etiologies.
To cite this abstract in AMA style:
Bell R, Brendel M, Xiang J, Consortium A, DiCarlo E, Anolik J, Donlin L, Orange D, Schwarz E, Ivashkiv L, Wang F. Machine Learning and Computational Pathology Can Reveal Synovial Pathotypes in Mice and Humans [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/machine-learning-and-computational-pathology-can-reveal-synovial-pathotypes-in-mice-and-humans/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/machine-learning-and-computational-pathology-can-reveal-synovial-pathotypes-in-mice-and-humans/