Session Information
Session Type: ACR Abstract Session
Session Time: 2:30PM-4:00PM
Background/Purpose: Different autoimmune diseases can co-exist in an individual and share similar genetic associations, autoimmune signaling pathways, and clinical manifestations. However, autoimmune diseases present varied cellular heterogeneities and may be distinguished by their primary target organs or tissues. The immune mechanisms that are shared between similar autoimmune diseases remain poorly understood due to limited access to affected human tissues and computational scalability. Recently, high resolution single-cell RNA-seq profiles have provided the opportunity for study of the contribution of diverse cell populations to disease pathogenesis. This advance has enabled unbiased comparison of disease across affected tissues with the goal of understanding autoimmune similarities.
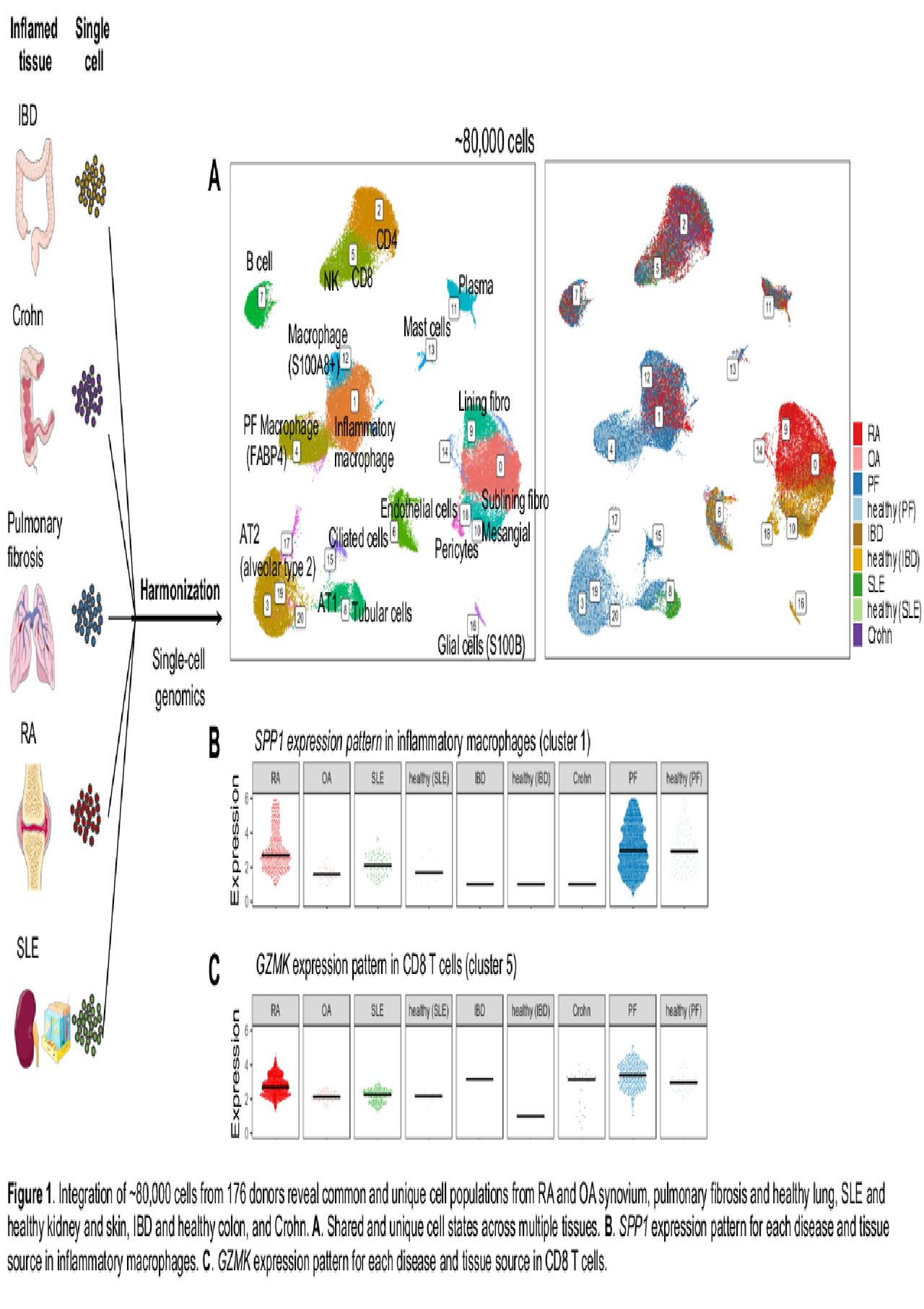
Methods: We have analyzed and integrated ~80,000 cells from 176 donors from publicly available single-cell RNA-seq datasets generated from rheumatoid arthritis (RA) and osteoarthritis (OA) synovium1, pulmonary fibrosis and healthy lung, systemic lupus erythematosus (SLE) and healthy kidney and skin2,3, inflammatory bowel disease (IBD) and healthy colon, and Crohn intestinal mucosal biopsies. We use a robust integrative strategy4 to cluster and project all the cells into two dimensional-space by correcting technical batch effect across tissue, donor, and single-cell platform (10X Genomics, Celseq2, Dropseq, Fluidigm, and Smartseq).
Results: We identify 21 diverse cell type populations across multiple tissue sources (Figure 1A). In the myeloid cell population, we observed four distinct subsets including VSIG4+ M2-like macrophages, S100A8+ macrophages, SPP1+ MMP9+ inflammatory macrophages, and dendritic cells (DC). The SPP1+ MMP9+ inflammatory macrophage population with high expression of matrix metalloproteinases genes is co-localized between macrophages in RA synovium, SLE kidney, and alveolar from pulmonary fibrosis lung, and is absent in healthy lung tissue (Figure 1B). For CD8 T cells and nature killer cells, we identified a shared transcriptional gradient of granzyme-expressing cytotoxic effectors between RA synovium, SLE kidney, and fibrotic lung. Interestingly, the GzmK+ CD8 T cell population is absent in the healthy lung (Figure 1C). The patterns of cytotoxic effector states may suggest similarities between the primary sites of inflamed RA and inflamed fibrotic lung, including potential common active pathways. In the stromal cell compartment, we observed distinct populations across different diseases and tissues, including fibroblasts, pericytes, mesangial, and tubular cells.
Conclusion: We demonstrate that integrative analyses between disease tissues by single-cell transcriptomics is capable to discovering shared and unique disease-specific gene expression modules and cell states, and may help predict potential therapeutic targets for inflammatory and fibrotic diseases.
Reference
- Zhang, F. et al. Nat Immunol (2019)
- Der, E., et al. Nat Immunol (2019)
- Arazi, A., et al. Nat Immunol, In press
- Korsunsky, I., et al. bioRxiv (2018)
To cite this abstract in AMA style:
Zhang F, Mears J, Korsunsky i, Wei K, Jonsson A, Rao D, Kim E, Donlin L, Buyon J, Petri M, Putterman C, Tuschl T, Hacohen N, Diamond B, Brenner M, Raychaudhuri S. Integration of Single Cells from Inflammatory Disease Tissues Reveals Common and Unique Pathogenic Cell States [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/integration-of-single-cells-from-inflammatory-disease-tissues-reveals-common-and-unique-pathogenic-cell-states/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/integration-of-single-cells-from-inflammatory-disease-tissues-reveals-common-and-unique-pathogenic-cell-states/