Session Information
Date: Sunday, November 7, 2021
Title: Abstracts: T Cell Biology & Targets in Autoimmune & Inflammatory Disease (0966–0969)
Session Type: Abstract Session
Session Time: 10:30AM-10:45AM
Background/Purpose: Cutaneous lupus erythematosus (CLE) is commonly present in patients with systemic lupus erythematosus (SLE), but can also exist as an isolated manifestation without further systemic involvement. Single cell RNA-sequencing (scRNAseq) has previously been employed to define the T cell states present in SLE and lupus nephritis (LN), which has helped reveal insights into the pathogenesis of the disease. Similar scRNAseq studies targeted at skin lesions in SLE could likewise help define the cellular landscape of cutaneous lupus, and could facilitate comparisons of cell infiltrates in different target tissues in SLE. Here, we present the scRNAseq profile of T and NK cell populations within both lesional and non-lesional skin biopsies of patients with cutaneous lupus. We further integrate this dataset of skin-localized T cells with scRNAseq data from T cells from LN kidneys to compare and contrast T cell infiltrates at these two target sites.
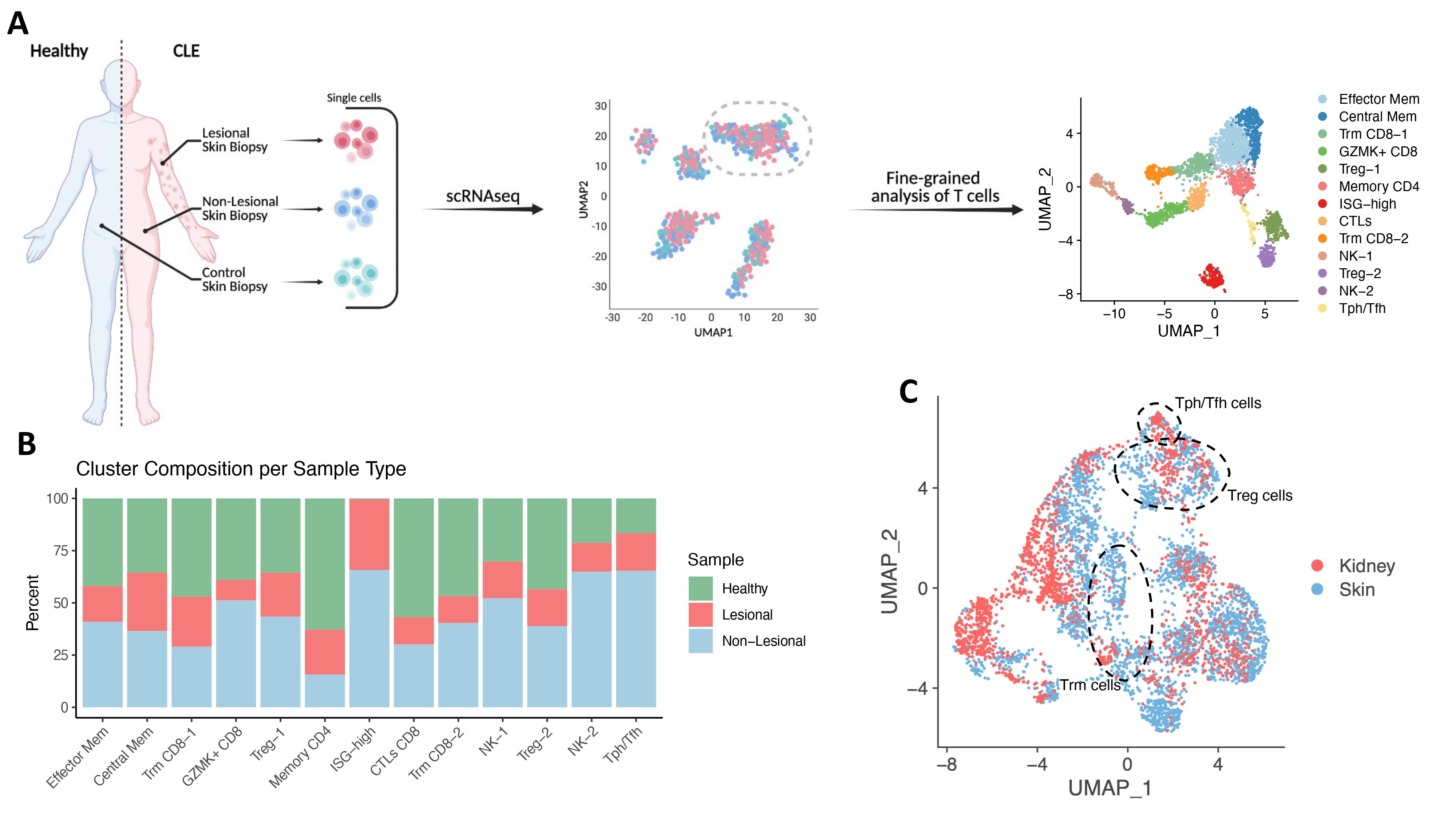
Methods: Skin biopsies were obtained from both lesional and non-lesional sites of patients with cutaneous lupus, and from healthy donors. Single cells were encapsulated and sequenced using the 10x Genomics Chromium platform, and data were processed using the Cell Ranger pipeline (10x Genomics). Broad T/NK cell clusters were subset from this dataset and were processed using Seurat for quality control filtering and subcluster analysis. scRNAseq data from LN T cells was obtained from the AMP RA/SLE Phase LN immune cell dataset. Integration of datasets was performed in Seurat using CCA analysis.
Results: Following filtering, paired lesional and non-lesional skin biopsies from 6 SLE patients and 1 isolated CLE patient, along with biopsies from 13 healthy donors, yielded 3,499 T and NK cells. Clustering of these cells revealed 13 subclusters. A population of strongly interferon-responding cells were present in patients with CLE but absent in healthy donors. Within CLE patients, both lesional and non-lesional cells across clusters expressed elevated levels of interferon simulated genes (ISGs), mirroring the ISG upregulation previously seen in scRNAseq studies of LN and blood T cells of lupus patients. Other populations enriched in lupus skin biopsies include a peripheral helper/follicular helper-like CD4 T cells (Tph/Tfh-like) population characterized by expression of CXCL13, MAF, and ICOS, and a population of NK cells expressing PRF1, GNLY, and GZMB. Integration of this dataset with T cells from LN samples using CCA revealed many shared T cell states, including populations of resident memory CD8 T cells, regulatory T cells, Tph/Tfh-like cells, cytotoxic CD8 T cells, ISG-high cells, and NK cells.
Conclusion: This data represents the first analysis of the transcriptomics of T and NK cells in cutaneous lupus at the single cell level. We defined the populations of T cells present in lesional skin as compared to non-lesional skin from the same patients, as well as skin from healthy controls. We identified a strong ISG upregulation in samples collected from CLE patients, and also noted an overrepresentation of helper T cells and NK cells among these samples. Integration of CLE and LN further revealed numerous conserved states, serving as a base for deeper analysis across tissues in lupus.
To cite this abstract in AMA style:
Dunlap G, Billi A, Ma F, Gudjonsson J, Kahlenberg J, Rao D. Integrated Single Cell RNA-Sequencing Analysis of Tissue-Localized T Cells in Cutaneous Lupus and Lupus Nephritis [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/integrated-single-cell-rna-sequencing-analysis-of-tissue-localized-t-cells-in-cutaneous-lupus-and-lupus-nephritis/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/integrated-single-cell-rna-sequencing-analysis-of-tissue-localized-t-cells-in-cutaneous-lupus-and-lupus-nephritis/