Session Information
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Mechanisms responsible for the distribution and severity of joint involvement in RA are not known. To explore whether site-specific FLS biology might contribute to location-specific synovitis and explain the predilection for hand (wrist/metacarpal phalangeal joints) in RA, we generated transcriptomic and chromatin accessibility data from FLS to define the transcription factors (TFs) and pathways unique to each location.
Methods: FLS were derived from hand, knee and hip surgical samples. Whole genome ATAC-seq and RNA-seq were obtained from 10 hand, 10 hip, and 10 knee FLS under unstimulated (unstim) or stimulated conditions (TNF [50 ng/ml for 6 h]). Differentially expressed genes (DEGs) were identified using the Wilcoxon test. TF motifs were queried within promoter/enhancer open chromatin. The assays were integrated using Taiji (Nat Commun 2022;13:6221) and Personalized PageRanks were quantified. Cell growth was assayed using MTT in medium and PDGF-stimulated FLS on day 7.
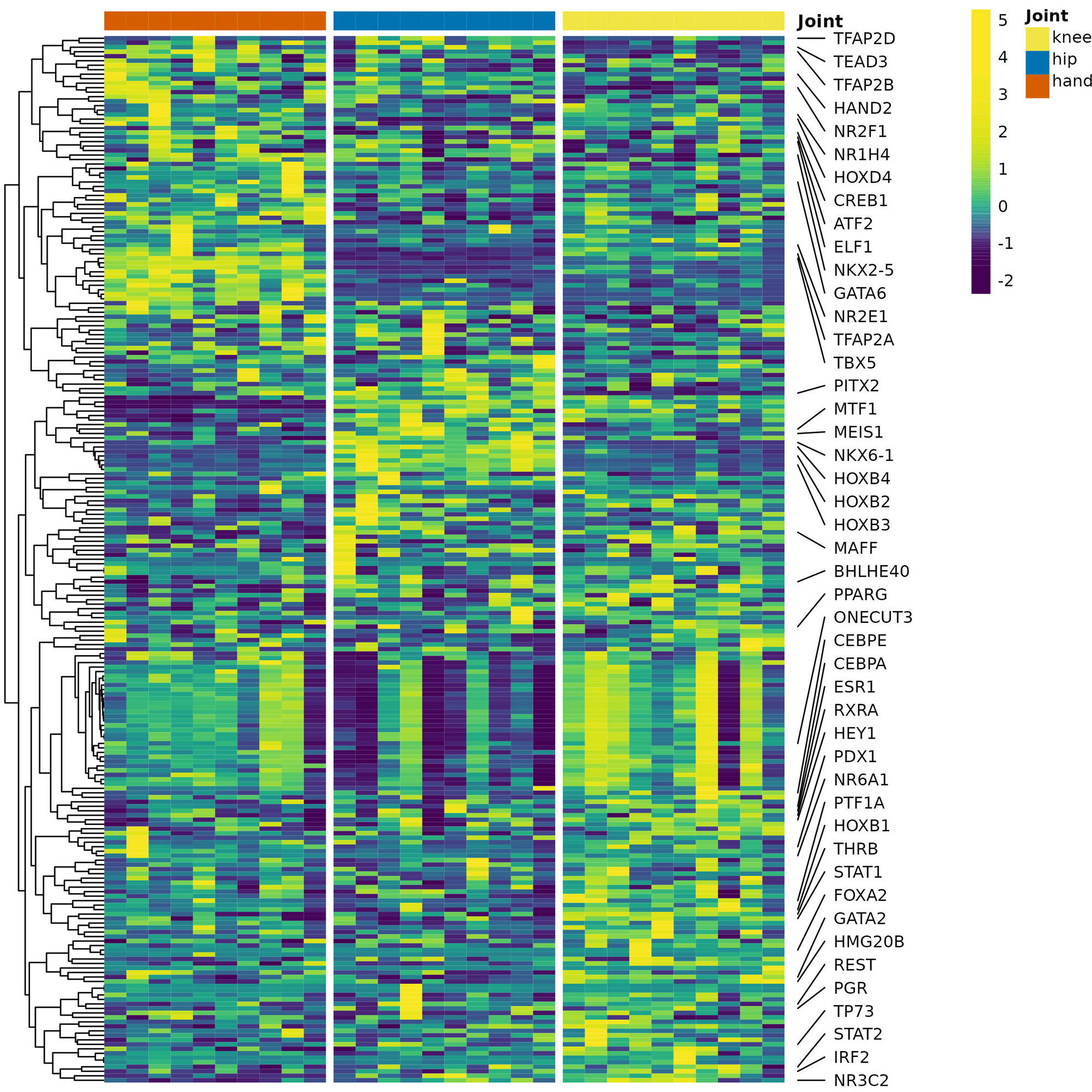
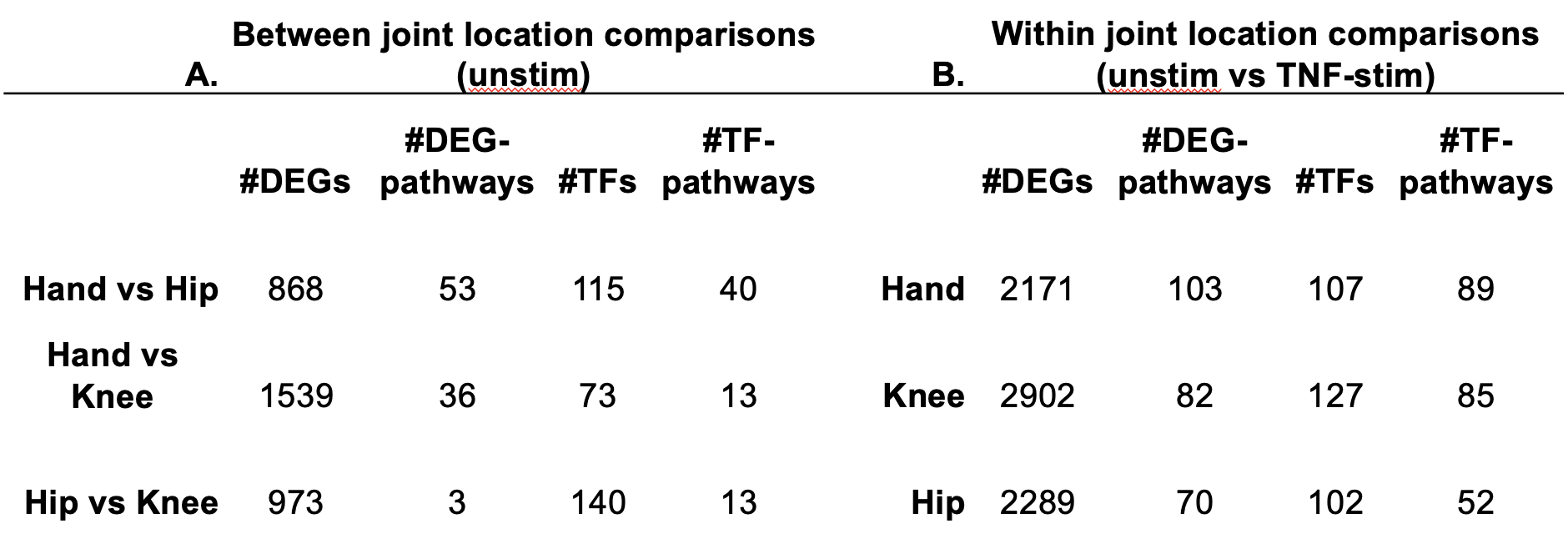
Results: Multiple DEGs between joints were identified in their respective FLS (Table 1A). Hand vs knee or hand vs hip FLS comparisons revealed increased expression of genes associated with proliferation/inflammation (eg, TLR2, VCAM1, MMP13) and decreased cell-cycle inhibitors (eg, CDKN1A, CDKN1C, CDKN2D) in hand. Pathway analysis revealed greatest enrichment for hand vs hip and included proinflammatory (eg, TNFs bind physiological receptors) and proliferative (eg, Integrin cell surface interactions) pathways (Table 1A). Taiji distinguished joint-specific TFs and identified greater PageRanks for key TFs (eg, ATF2, CREB1) in hand (Fig 1, Table 1A). Pathway analysis revealed greatest enrichment for hand vs hip and included proliferation/inflammation pathways (eg, MAPK activation, PIP3 activates AKT signaling) (Table 1A). The predicted differences in proliferation were confirmed in a cell growth assay showing that hand FLS proliferation was greater than hip or knee after PDGF-stim (Hand/knee or hip ratio=1.4; P< 0.001). The joint-specific transcriptome differences for hand vs hip and knee were enhanced by TNF-stim. DEGs within a particular joint between unstim vs TNF-stim revealed greatest pathway enrichment in hand (Table 1B). TNF-stim hand FLS had unique induction of proliferation/inflammation genes (eg, TANK, KSR1, CDC14A) and pathways (eg, Diseases of signal transduction by growth factor receptors/second messengers, Regulation of necroptotic cell death), and these pathways were not observed in TNF-stim hip or knee. TNF-stim in hand FLS had increased PageRanks of key inflammatory TFs (eg, RELA, STAT3) and pathways (eg, MAPK activation, Signaling by ALK in cancer) that were not observed for hip/knee.
Conclusion: Integrated analysis of FLS identified joint-specific patterns of gene expression and TFs, including pathway enrichment in hand FLS associated with inflammation and proliferation. These differences are more prominent after TNF stimulation. Hand joints are commonly earlier and are more severe in RA. Distinctive joint-specific FLS biology associated with hand-specific pathways might explain the distribution and severity of joints involved in RA.
To cite this abstract in AMA style:
Choi E, R. L. Machado C, Boyle D, Wang W, Firestein G. Integrated Analysis of Rheumatoid Arthritis (RA) Fibroblast-like Synoviocytes (FLS) Transcriptome and Chromatin Accessibility Identifies Mechanisms Associated with Location-specific Disease Severity [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/integrated-analysis-of-rheumatoid-arthritis-ra-fibroblast-like-synoviocytes-fls-transcriptome-and-chromatin-accessibility-identifies-mechanisms-associated-with-location-specific-disease-severity/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/integrated-analysis-of-rheumatoid-arthritis-ra-fibroblast-like-synoviocytes-fls-transcriptome-and-chromatin-accessibility-identifies-mechanisms-associated-with-location-specific-disease-severity/