Session Information
Date: Saturday, November 12, 2022
Title: Abstracts: Miscellaneous Rheumatic and Inflammatory Diseases I
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Systemic immune-mediated diseases are heterogeneous, and the pathogenesis varies among and within each disease. Some studies tried to stratify patients with immune-mediated diseases into some subgroups by the data of molecular patter or mass spectrometry [1,2]; however, to date, classification of the disease generally is based on symptoms and some specific serological features, and the treatment is relatively uniform depending on the diagnosis and affected organs. We conducted this study to reveal what kinds of immune cells are involved in each disease and reclassify them with cell proportions.
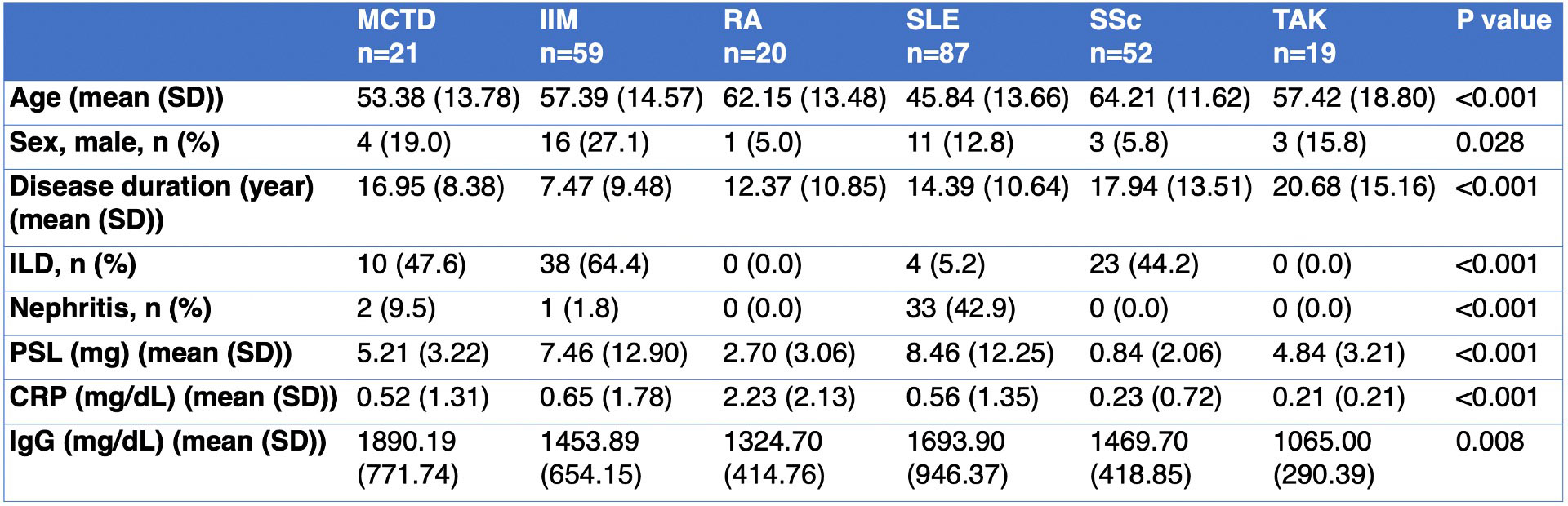
Methods: Immunophenotyping of 258 patients with systemic immune-mediated diseases, including 21 mixed connective tissue disease (MCTD), 59 idiopathic inflammatory myopathy (IIM), 20 rheumatoid arthritis (RA), 87 systemic lupus erythematosus (SLE), 52 systemic sclerosis (SSc), and 19 Takayasu arteritis (TAK) were undertaken (Table 1). We collected the clinical data and sorted a total of 32 types of immune cells from PBMCs using flow cytometry. We selected 11 out of 32 disease-specific immune cells whose variances were more than 0.10 and compared the proportion of immune cells and clinical data in each population. Categorical and continuous variables were compared using the chi-square test and analysis of variance (ANOVA). A hierarchical clustering procedure by calculating Euclidean distance and using Ward’s method was performed to form clusters from the Z score of immune cell proportions.
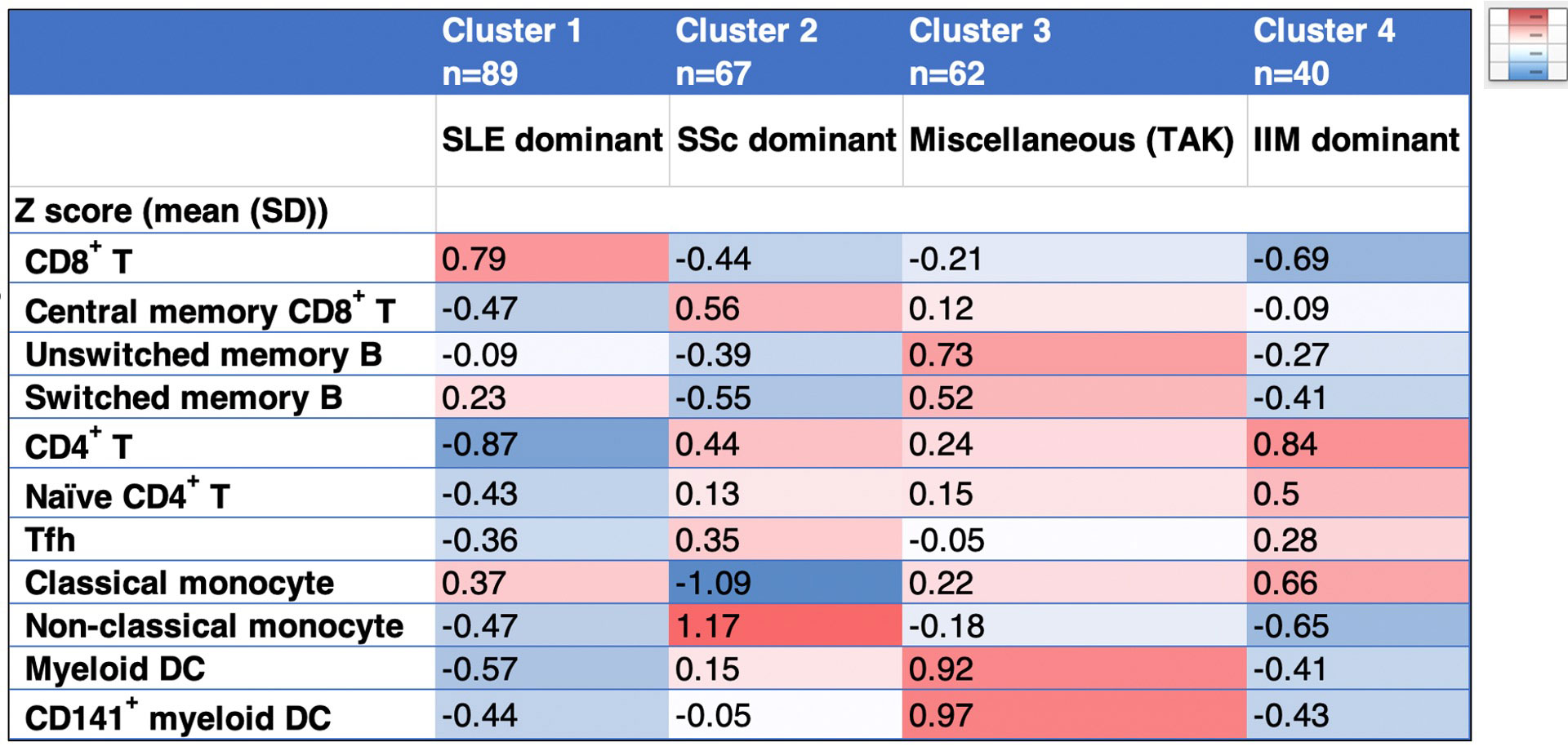
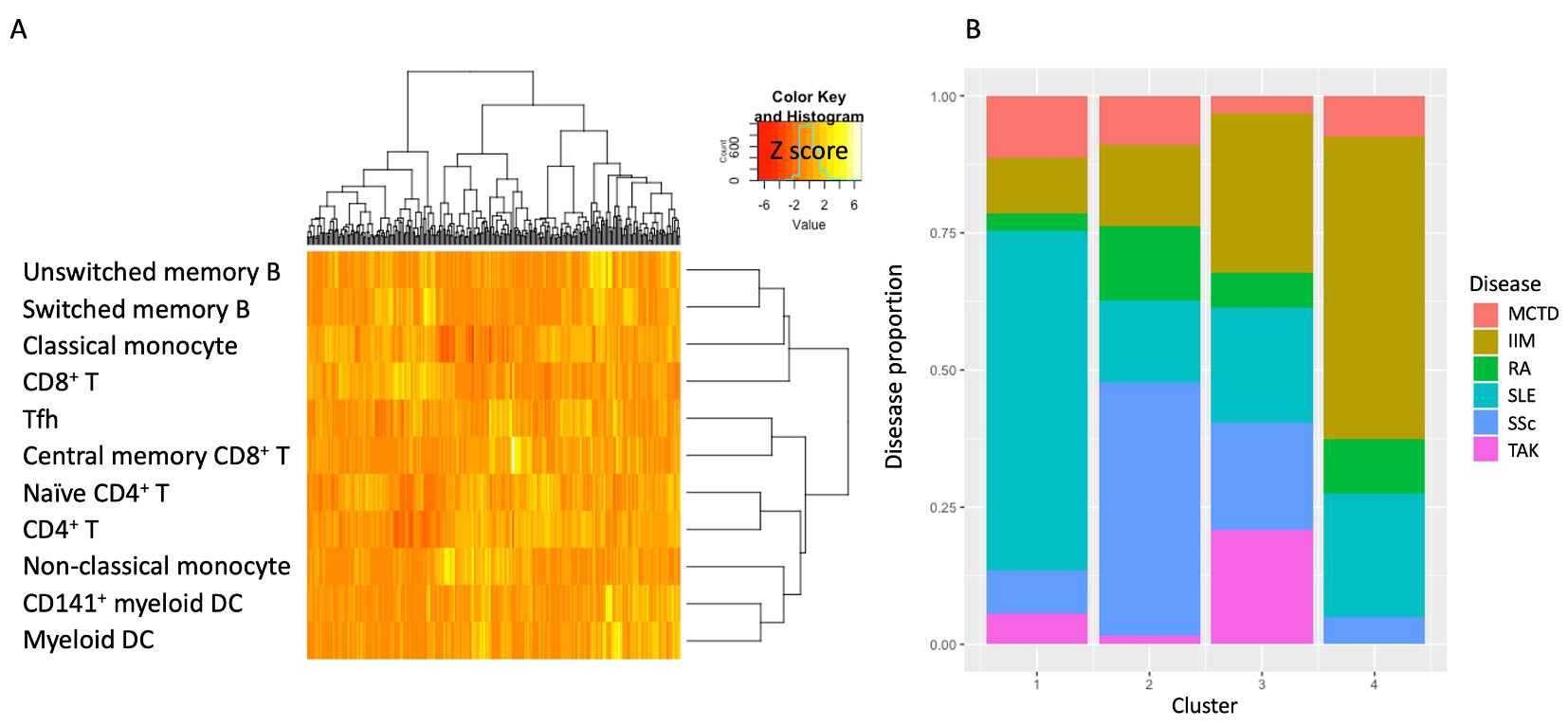
Results: The hierarchical clustering of flow cytometry data of 11 immune cells, including CD8+ T, central memory CD8+ T, CD4+ T, naïve CD4+ T, follicular helper T (Tfh), switched memory B, unswitched memory B, classical monocytes, non-classical monocytes, myeloid dendritic cell (DC), and CD141+ myeloid DC identified 4 clusters (Table 2, Figure 1A). Cluster 1,2, and 4 were dominant SLE, SSc, and IIM, respectively (Figure 1B). Cluster 3 contained miscellaneous diseases but most TAK patients were classified into this group. Each cluster showed high proportions of CD8+ T in cluster 1, central memory CD8+ T, Tfh, non-classical monocytes in cluster 2, unswitched memory B, switched memory B, myeloid DC, and CD141+ myeloid DC in cluster 3, and CD4+ T, naïve CD4+ T, and classical monocytes in cluster 4.
Conclusion: The systemic immune-mediated diseases such as MCTD, IIM, RA, SLE, SSc, and TAK were classified into four groups by the immune cell proportions. Our data may corroborate the understanding of the pathogenesis of various systemic immune-mediated diseases and can be a clue to reclassifying them by the associated immune cells.
To cite this abstract in AMA style:
Izuka S, Komai T, Itamiya T, Ota M, Yamada S, Nagafuchi Y, Shoda H, Matsuki K, Yamamoto K, Okamura T, Fujio K. Immunophenotypic Categorization of Systemic Immune-mediated Diseases [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/immunophenotypic-categorization-of-systemic-immune-mediated-diseases/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/immunophenotypic-categorization-of-systemic-immune-mediated-diseases/