Session Information
Session Type: Abstract Session
Session Time: 1:00PM-2:30PM
Background/Purpose: The largest genetic risk factor for RA localizes to the MHC Class II HLA-DRB1 gene, which encodes the machinery for antigen presentation to CD4 T cells. The mechanisms by which CD4 T cells may be involved in this elevated risk is unclear. To elucidate potential pathogenic mechanisms, here we further explored the recent demonstration by Ishigaki et al. (2022) that in healthy individuals, the presence of high-risk RA-associated HLA-DRB1 alleles results in thymic selection of specific patterns of T cell receptors (TCRs) in the CD4 T cell repertoire, and that those patterns are enriched among CD4 T cell clones that recognize citrullinated antigens.
Methods: We obtained synovial tissue (n=15) and blood (n=17) from patients (n=19), among which 13 were paired, satisfying either the 1987 or 2010 ACR/EULAR RA classification criteria, with informed consent and IRB approval. We isolated CD4 T cells and performed single-cell transcriptome and TCR sequencing (10X platform) with yield of ~5,000-9,000 cells per sample. We applied a custom computational pipeline to isolate high-quality single-cell data, infer HLA-DRB1 genotypes, and quantify TCR repertoire metrics including clonal expansion and the Ishigaki et al. TCRβ CDR3 risk score. With this dataset and public CD4 TCR and HLA data of healthy individuals’ blood (Emerson et al. 2017), we also computed associations between V and J gene usage and HLA-DRB1 genotype.
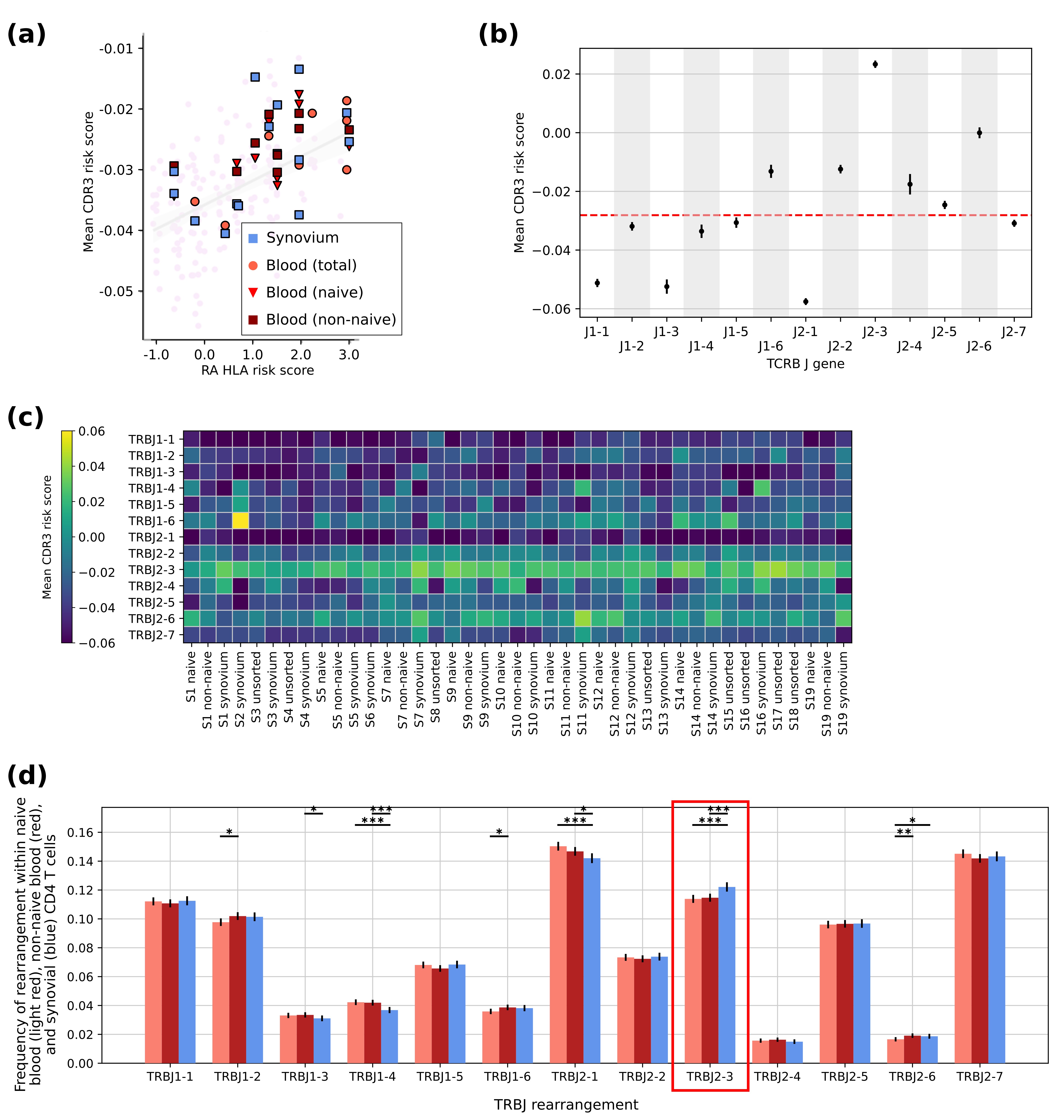
Results: Here we identified CD4 TCR clones harboring TCRβ CDR3 features selected by HLA-DRB1 risk alleles in RA patients, in increasing abundance with increasing risky HLA allele burden (Fig 1a). In an effort to define additional features of clones with features selected by the high-risk HLA-DRB1 alleles, we noted a highly significant enrichment of clones containing the TRBJ2-3 gene rearrangement (Fig 1b), which is consistent across all RA patients (Fig 1c). Examining the portion of the CDR3 regions overlapping with the germline-encoded TRBJ2-3 gene, we detected negatively charged TCR amino acids that are predicted to interact with the HLA-DRB1 risk-encoded amino acids. In the synovium of RA patients, we demonstrate that across nearly one hundred thousand CD4 T cells, the size of each CD4 T cell clone is small, below 1% of the total CD4 population. Nonetheless, in comparison to all other TRBJ genes, we detected enrichment of the TRBJ2-3 CD4 T cell clones in RA synovium (Fig 1d).
Conclusion: Our study has defined TCR receptor repertoire patterns that associate with high-risk HLA-DRB1 alleles in RA patients, and identified the CD4 T cell subset with TRBJ2-3 as containing a particularly high concentration of those patterns. Given the small clone sizes for CD4 T cells in general, and that TRBJ2-3-containing CD4 clones are enriched across the CD4 T cell repertoire in individuals with risky HLA-DRB1 alleles, we argue that CD4 T cell involvement in RA synovial pathology may be greatly influenced by germline-encoded features distributed across the CD4 T cell repertoire. These findings may motivate therapeutic targeting of CD4 T cell subsets in RA patients that are defined by their germline-encoded regions, including TRBJ2-3.
To cite this abstract in AMA style:
Lakhanpal A, Ishigaki K, Singaraju A, Kochen A, Fein M, Raychaudhuri S, Donlin L. HLA-DRB1 Rheumatoid Arthritis (RA) Risk Alleles Preferentially Select TRBJ2-3-containing CD4 T Cells in RA Patients [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/hla-drb1-rheumatoid-arthritis-ra-risk-alleles-preferentially-select-trbj2-3-containing-cd4-t-cells-in-ra-patients/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/hla-drb1-rheumatoid-arthritis-ra-risk-alleles-preferentially-select-trbj2-3-containing-cd4-t-cells-in-ra-patients/