Session Information
Date: Friday, November 6, 2020
Title: Miscellaneous Rheumatic & Inflammatory Diseases I: Mechanisms of Disease (0459–0463)
Session Type: Abstract Session
Session Time: 10:00AM-10:50AM
Background/Purpose: Rheumatoid arthritis (RA)-associated inflammatory lung disease is an extra-articular manifestation of RA associated with increased morbidity and mortality, whose precise molecular mechanisms remain undetermined. We established a novel mouse model of combining the collagen-induced arthritis (CIA) model with the organic dust extract (ODE)-induced airway inflammatory disease model to demonstrate that combined exposures (CIA+ODE) increased arthritis, autoantibody levels, and polarized the lung towards pre-fibrotic inflammatory processes. Utilizing this animal model, we aimed to identify relevant cellular and protein mediators through high-throughput single-cell analysis.
Methods: Arthritis-prone DBA/1J mice were assigned to 1 of 4 treatment groups: Sham, CIA, ODE, and CIA+ODE for 5 weeks. Whole lung cells were processed for single cell RNA-sequencing (scRNA-seq). In separate studies, 4 defined myeloid-derived immune cell populations consisting of macrophages, monocytes/macrophages, monocytes, and neutrophils were isolated from whole lung of exposed mice by fluorescence activated cell sorting (FACS) with gene expression determined by NanoString (771 genes). Data are represented as fold-change compared to Sham from 3 experiments (2-3 mice/group/experiment) using two-way ANOVA.
Results: By scRNA-seq, 14 discrete lung immune cell populations were identified through unsupervised clustering and respective gene expression. Cell populations included 3 neutrophils (inflammatory, resident/transitional, autoreactive/myeloid derived suppressor), 5 macrophages (airspace, differentiating/recruited, recruited, resident/interstitial, and proliferative airspace), 2 T-cells (differentiating and effector), and a population of inflammatory monocytes, dendritic cells, B-cells and natural killer cells (Figure 1). Inflammatory monocytes, autoreactive/suppressor neutrophils, and recruited/differentiating macrophage subpopulations were predominantly ascribed to arthritis induction (CIA and CIA+ODE). Genes highly expressed with arthritis induction across lung cell populations of myeloid origin included interferon-related, autoimmunity, and suppressive genes (e.g. Ifit3b, Ifit3, S100a8, and S100a9). NanoString analysis of the 4 isolated lung myeloid-derived populations were consistent with scRNA-seq findings (Figure 2,3). Several immunosuppressive and autoimmune genes (e.g. Oasl1, Oas2, Ifit3, Gbp2, Ifi44, Ifi44l, Zbp1, and Ifit1) were disproportionately expressed in lung cell populations of neutrophils and macrophages/monocytes among CIA and CIA+ODE groups. Complement cascade genes (e.g. C1s1, Cfb, C1qa, and C1ra) were uniquely increased in the co-exposure (CIA+ODE) across all 4 cell populations.
Conclusion: Recruited and inflammatory macrophages/monocytes and neutrophils expressing suppressive and autoimmune genes in systemic autoimmunity might contribute towards pro-fibrotic inflammatory responses in the lung following airborne biohazard exposures. Targeting these cellular features and/or signature genes could lead to novel approaches in RA-associated inflammatory lung disease.
 Figure 1. Single-cell RNA-sequencing analysis. Top left panel shows unsupervised gene clustering of all CD45+ gene expressing lung cells when all treatment groups are aggregated. Cell names are ascribed based upon gene expression (bottom panel). Top right panel depicts clustering of the defined aggregated clustering across treatment groups with dotted circles highlighting macrophages, monocytes, and granulocytes. Arbitrary numbers (1, 2, etc.) designate unique gene clusters determined by unsupervised clustering.
Figure 1. Single-cell RNA-sequencing analysis. Top left panel shows unsupervised gene clustering of all CD45+ gene expressing lung cells when all treatment groups are aggregated. Cell names are ascribed based upon gene expression (bottom panel). Top right panel depicts clustering of the defined aggregated clustering across treatment groups with dotted circles highlighting macrophages, monocytes, and granulocytes. Arbitrary numbers (1, 2, etc.) designate unique gene clusters determined by unsupervised clustering.
 Figure 2. Lung cell populations of macrophages (MΦ) and monocytes (Mo) isolated by FACS and gene expression analysis among treatment groups. A, Representative dot plots of macrophages (CD11chigh CD11bvariable), monocytes-macrophages (CD11cintermediate CD11bhigh), and monocytes (CD11c– CD11bhigh) from each treatment of Sham, CIA, ODE, and CIA+ODE shown. B, C, and D Heatmaps of each cell population depicting the fold-change of top 5 genes/treatment group (CIA, ODE and CIA+ODE), normalized to 20 housekeeping genes and compared to Sham.
Figure 2. Lung cell populations of macrophages (MΦ) and monocytes (Mo) isolated by FACS and gene expression analysis among treatment groups. A, Representative dot plots of macrophages (CD11chigh CD11bvariable), monocytes-macrophages (CD11cintermediate CD11bhigh), and monocytes (CD11c– CD11bhigh) from each treatment of Sham, CIA, ODE, and CIA+ODE shown. B, C, and D Heatmaps of each cell population depicting the fold-change of top 5 genes/treatment group (CIA, ODE and CIA+ODE), normalized to 20 housekeeping genes and compared to Sham.
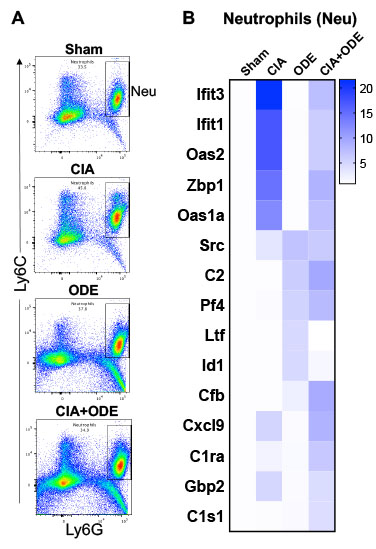 Figure 3. Lung cell populations of neutrophils (Neu) isolated by FACS and gene expression analysis among treatment groups. A, Representative dot plots of neutrophils (Ly6C+ Ly6G+) from each treatment of Sham, CIA, ODE, and CIA+ODE shown. B, Heatmap depicting the fold-change of top 5 genes/treatment group (CIA, ODE and CIA+ODE), normalized to 20 housekeeping genes and compared to Sham.
Figure 3. Lung cell populations of neutrophils (Neu) isolated by FACS and gene expression analysis among treatment groups. A, Representative dot plots of neutrophils (Ly6C+ Ly6G+) from each treatment of Sham, CIA, ODE, and CIA+ODE shown. B, Heatmap depicting the fold-change of top 5 genes/treatment group (CIA, ODE and CIA+ODE), normalized to 20 housekeeping genes and compared to Sham.
To cite this abstract in AMA style:
Gaurav R, Mikuls T, Thiele G, Nelson A, Niu M, Guda C, Eudy J, Barry A, Romberger D, Duryee M, England B, Poole J. High-Throughput Single-Cell Analysis Reveals Unique Lung Cellular Subsets in a Murine Model of Rheumatoid Arthritis-Inflammatory Lung Disease [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/high-throughput-single-cell-analysis-reveals-unique-lung-cellular-subsets-in-a-murine-model-of-rheumatoid-arthritis-inflammatory-lung-disease/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/high-throughput-single-cell-analysis-reveals-unique-lung-cellular-subsets-in-a-murine-model-of-rheumatoid-arthritis-inflammatory-lung-disease/
