Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Lupus nephritis (LN) treatment decisions are typically informed using histopathological classification based on the International Society of Nephrology/Renal Pathology Society (ISN/RPS) and NIH activity and chronicity indices. LN class and activity can change over time, which often requires changes in treatment. However, serial kidney biopsies are invasive and impractical; thus, noninvasive biomarkers of LN classification are needed to help guide treatment decisions. Here, we investigated serum proteomic profiles to define noninvasive biomarkers of histological class and activity and chronicity indices.
Methods: SLE patients with LN (n=184) were recruited for this study as part of the Accelerating Medicines Partnership RA/SLE network. Kidney biopsies were evaluated by a renal pathologist according to ISN/RPS classification and NIH activity and chronicity indices. Serum samples were collected at the time of kidney biopsy, and protein expression was measured using the Olink Explore HT. Multivariate logistic regression, adjusting for age, gender, and genetic ancestry, and random forest algorithms were used to identify biomarkers of LN class and activity and chronicity indices.
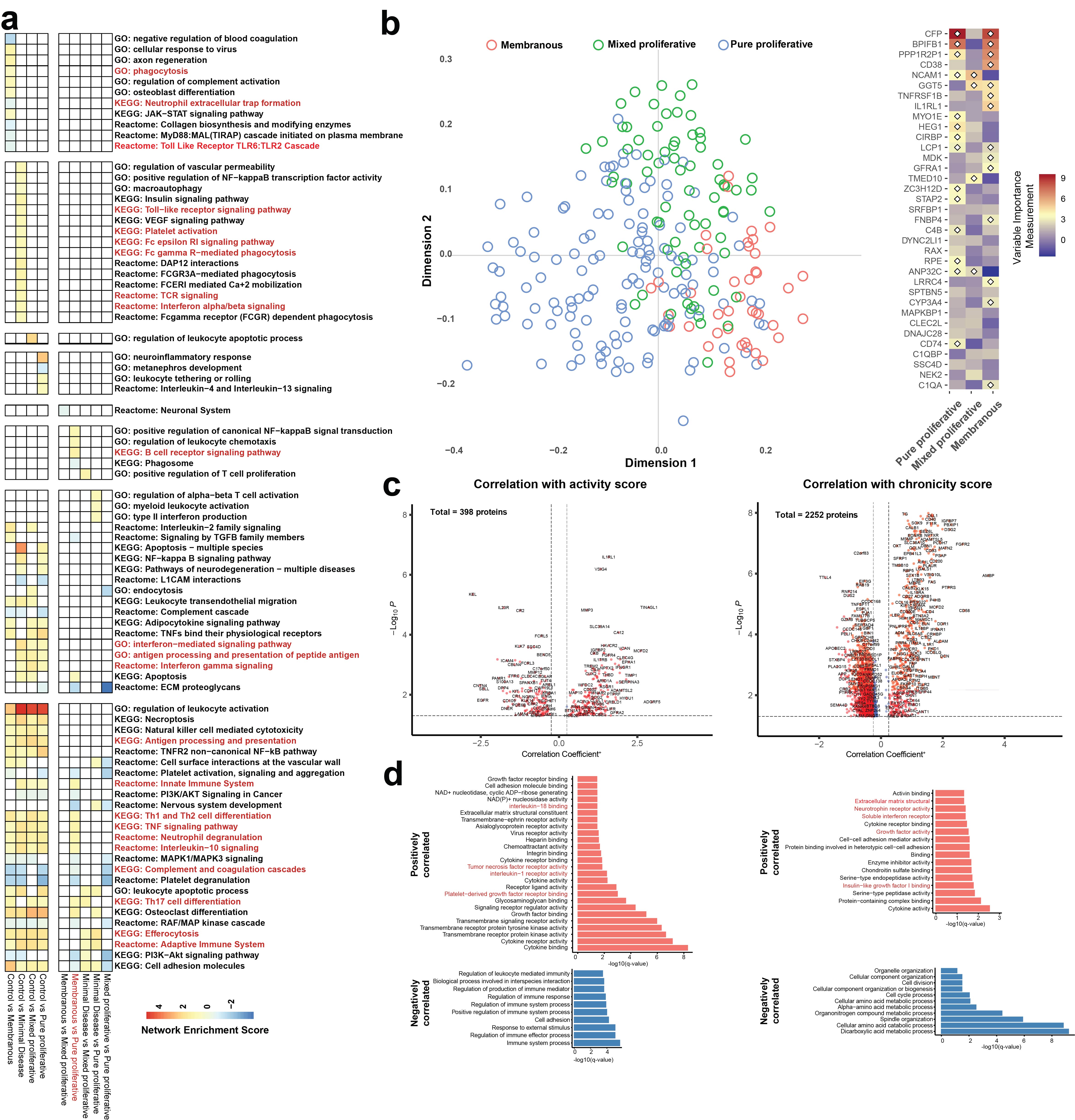
Results: Compared to healthy controls, LN patients upregulated multiple pathways related to the innate and adaptive immune systems, including TNF, IL-10, efferocytosis, and antigen processing and presentation pathways (Figure 1A). Patients with pure proliferative LN (class III/IV) showed further upregulation in B cell receptor signaling, Th1/Th2 differentiation, neutrophil degranulation, Th17 differentiation, and leukocyte chemotaxis pathways compared to those with minimal disease (class I/II), membranous (V), or mixed proliferative (III/IV+V) LN (Figure 1A). Machine learning models achieved high accuracy for distinguishing healthy controls (95.3% [86.9%-99%]) and LN patients (99.5%, [97% – 100%]) but were only able to distinguish pure proliferative (84.5% [75.8%, 91.1%]) from the other LN subclasses (Figure 1B). In addition, the expression of 398 and 2252 proteins was associated with the NIH activity and chronicity indices, respectively (Figure 1C). Specifically, proteins involved in IL-18, TNF, and IL-1 pathways and intracellular proteins from multiple organ systems with prominent enrichment in immune cells positively correlated with the activity index (Figure 1D). Interestingly, proteins enriched in interferon, growth factor, and neurotrophin receptor pathways and intracellular proteins from multiple organ systems, particularly the nervous system, correlated with the chronicity index (Figure 1D).
Conclusion: Serum protein expression pathways may serve as noninvasive biomarkers of LN class and activity and chronicity indices, helping to direct therapeutic decisions.
To cite this abstract in AMA style:
Lu R, Fava A, Jones B, Izmirly P, Anolik J, Putterman C, Wofsy D, Kretzler M, Berthier C, Woodle E, Weisman M, Ishimori M, Accelerating medicines Partnership: RA/SLE Network T, Diamond B, Buyon J, Petri M, Furie R, James J, Guthridge J. High-Throughput Proteomic Profiling of Sera as a Non-Invasive Method for Identifying Lupus Nephritis Subtypes [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/high-throughput-proteomic-profiling-of-sera-as-a-non-invasive-method-for-identifying-lupus-nephritis-subtypes/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/high-throughput-proteomic-profiling-of-sera-as-a-non-invasive-method-for-identifying-lupus-nephritis-subtypes/