Session Information
Date: Saturday, November 16, 2024
Title: Abstracts: T Cell Biology & Targets in Autoimmune & Inflammatory Disease
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Multiple T cell subsets, including Th1, Th17 cells, and Tfh/Tph, contribute to rheumatoid arthritis (RA) pathology. Clinical trials have shown efficacy of T cell co-stimulation modulation with abatacept in delaying clinical RA onset, indicating T cell involvement prior to clinical RA. However, the mechanisms by which T cell subsets contribute to progression to disease remain incompletely understood. In this study, we investigated 1) the CD4 T cell activation program in a group of ACPA+ individuals who later convert to clinical disease and 2) the epigenetic and transcriptomic profiles in CD4 naïve cells prior to clinical RA.
Methods: We conducted scRNA-seq profiling of peripheral blood mononuclear cells (PBMCs) from 16 ACPA+ individuals (Converters) over a period of up to 2 years before they developed clinical RA. Additionally, we performed simultaneous trimodal single cell profiling in ACPA+ at-risk individuals (ARI, n=7) and ACPA- controls (Controls, n=5). Non-negative matrix factorization (NMF) was used to project our data to a human CD4 T cell gene program reference (Yasumizu et al. 2024), followed by Leiden clustering. Cluster frequency was centered log-ratio transformed (CLR) then evaluated by linear mixed models. In multimodal data, differential chromatin accessibility was evaluated using MOCHA and transcription factor activities were inferred by ChromVAR.
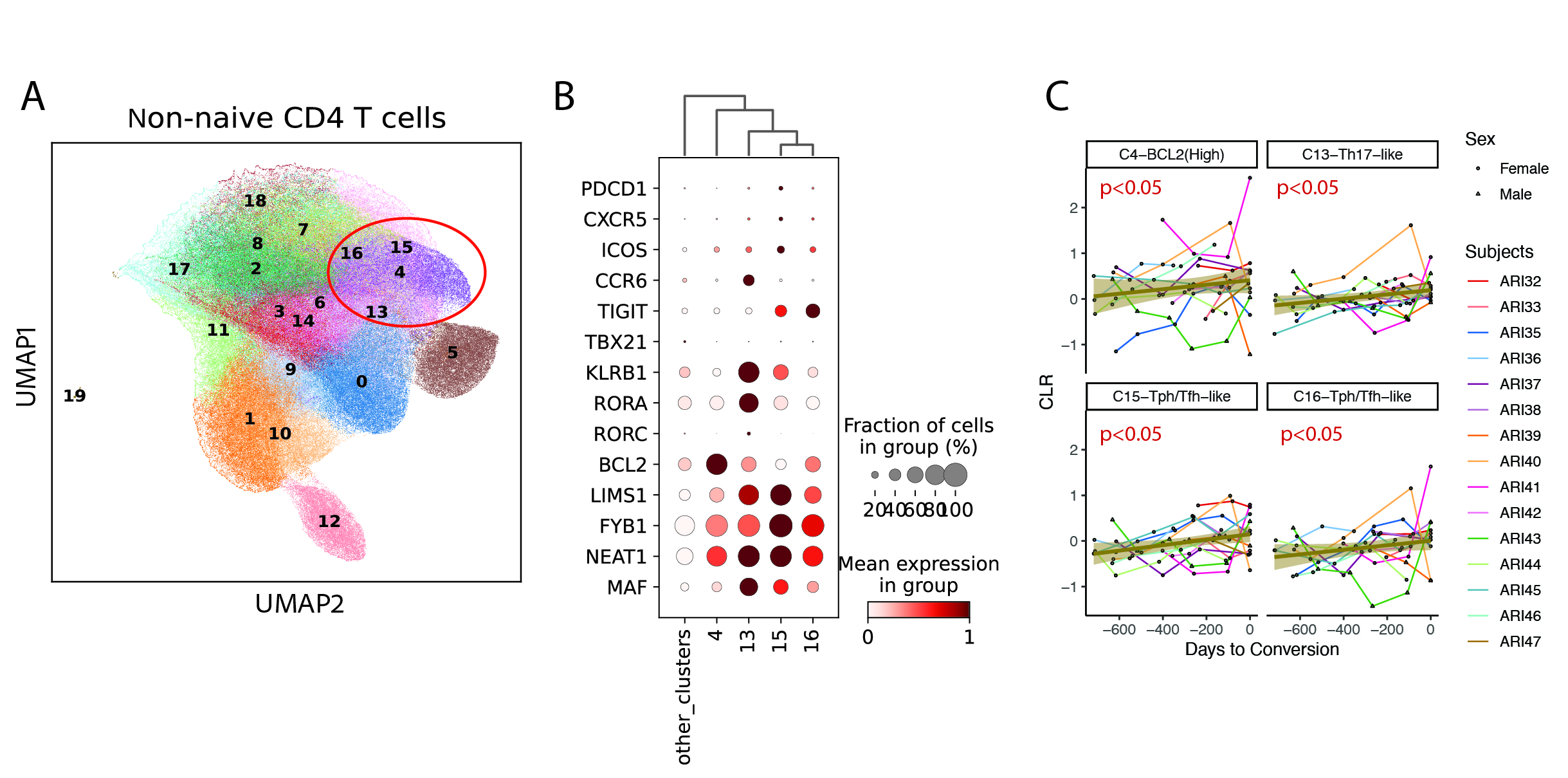
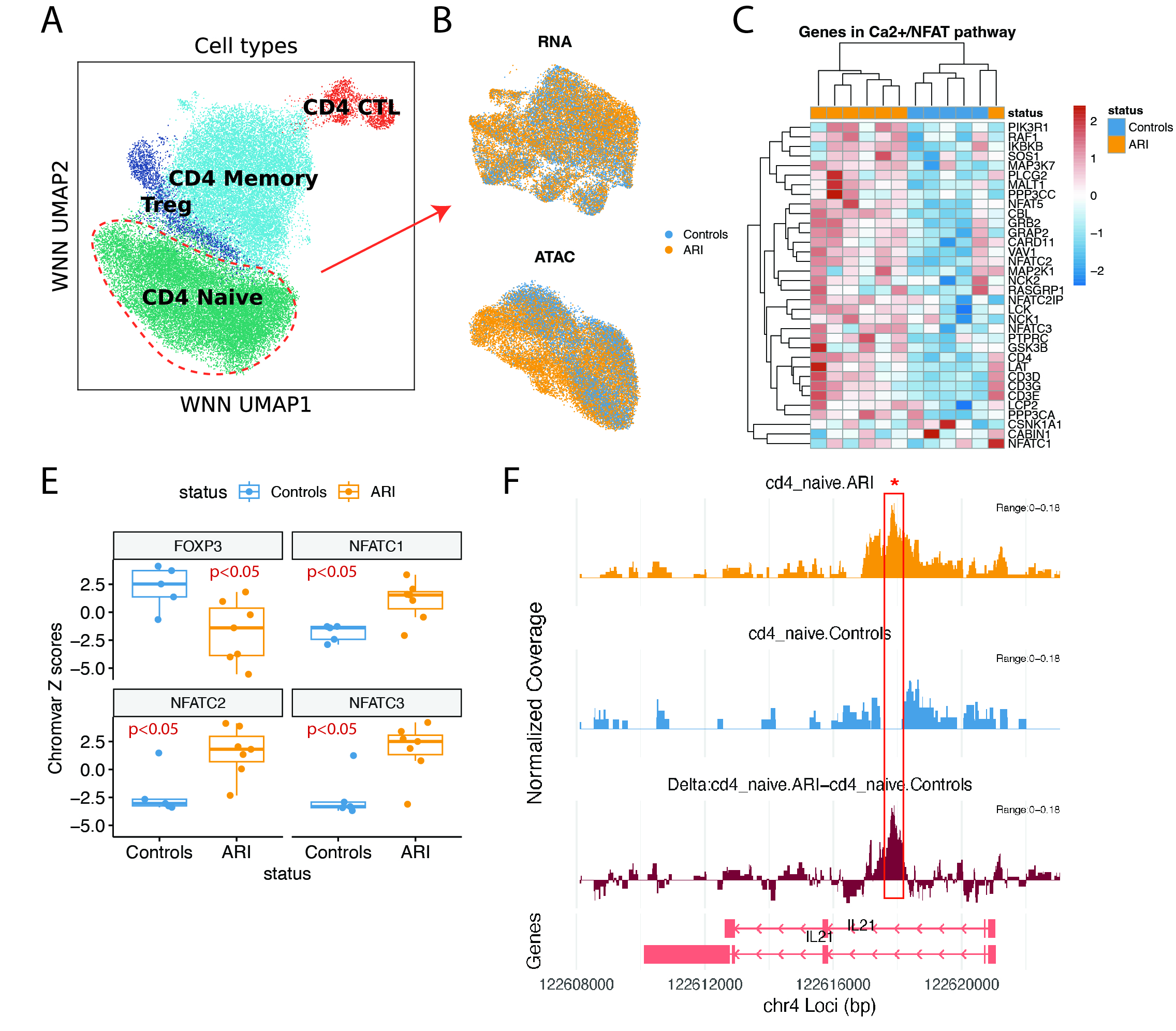
Results: In converters, we identified 19 non-naïve CD4 T cell clusters, of which 4 showed increased frequencies (p< 0.05) during progression to clinical RA. These clusters included: PDCD1+CXCR5+ ICOS+ Tph/Tfh-like C15 and C16; CCR6+KLRB1+RORA+ C13 Th17-like; BCL2high C4, suggesting a bias towards Tph/Tfh and Th17 phenotypes among these cells. Furthermore, these 4 clusters shared a common gene signature enriched for LIMS1, FYB1, NEAT1, MAF. (Fig 1) By integrating the surface protein, transcriptomic and epigenomic data, we found significant enrichment of NFATs, less FOXP3 motif (p< 0.05), and upregulation of calcium–calcineurin related gene expression in ARI compared to HC. Moreover, we discovered an IL-21 enhancer region in CD4 naive cells specifically enriched in ARI (p.adj< 0.01). (Fig 2) Notably, this region was enriched in the naive and Tfh cells compared with other T cell subsets in a publicly available human tonsil dataset (King et al 2021), suggesting an epigenetic imprint in the naïve cells connected to Tfh development.
Conclusion: Our results showed that CD4 T effector programs in converters were biased towards Tfh/Tph and Th17 development during progression to clinical RA, with a shared underlying gene signature suggesting a common activation process may lead to the development of phenotypically distinct CD4 effector cells in RA development. Integrative cross-sectional analyses in APCA+ ARI indicated enhanced TCR signaling occurred in naïve cells with an epigenetic imprint linked to Tfh development. Overall, our results suggest that epigenetic reprogramming of naïve cells contributes to the dysregulation of T cell activation in ACPA+ individuals prior to development of clinical RA, and provide mechanistic evidence for T cell targeting in an at-risk period for preventing or delaying clinical RA.
(A) UMAP of non-naive CD4 T cells clustered using NMF factors on gene programs identified as discriminating by Yasumizu et al. (B) Dot plot of the key marker genes used to identify the specific T helper phenotypes. Circle color indicates mean transcript detection; circles are sized proportional to the number of cells with transcript detection. (C) Line plot of centered log-ratio (CLR) transformed frequency changes as converters progress to clinical disease. Each participant’s longitudinal series is connected by a line with the group trendline shown (heavy yellow line). P-values indicated the association of CLR with time to disease conversion in a linear mixed model adjusted for age, sex, and BMI.
(A) Three-way nearest neighbor UMAP of CD4 T cells in the trimodal dataset. (B) UMAP of CD4 naive cells based on RNA and ATAC modalities, colored by cells from ARI or Controls. (D) ChromVAR Z-scores of NFAT family and FOXP3 inferred by chromatin accessibility. Wilcoxon tests were used to compare Z-scores between ARI and Controls, pseudobulked by samples. (E) Heatmap of scaled gene expression of selected genes in the NFAT-calcium–calcineurin pathway. (F) Gene track plot of IL21 gene accessibility in CD4 naive cells in ARI, Controls, and the delta between ARI and Controls. The red box highlights the 500 base pair peak that is significantly different, with adjusted p-values < 0.01.
To cite this abstract in AMA style:
He Z, Venkatesan P, Savage A, Glass M, Okada L, Krishnan U, Bennett C, Tran N, He Y, Rachid Zaim S, Ravisankar P, Garber J, Genge P, Lee K, Mettey R, Phalen C, Khosa S, Feser M, Zhang F, Boyle D, Kuhn K, Demoruelle K, Speake C, Buckner J, Goldrath A, Bumol T, Holers V, Skene P, Firestein G, Li X, Deane K, Torgerson T, Gillespie M. Epigenetic Control of Pathogenic CD4 T Cell Polarization During Progression to Rheumatoid Arthritis (RA) [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/epigenetic-control-of-pathogenic-cd4-t-cell-polarization-during-progression-to-rheumatoid-arthritis-ra/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/epigenetic-control-of-pathogenic-cd4-t-cell-polarization-during-progression-to-rheumatoid-arthritis-ra/