Session Information
Date: Saturday, November 6, 2021
Title: SLE – Diagnosis, Manifestations, & Outcomes Poster I: Diagnosis (0323–0356)
Session Type: Poster Session A
Session Time: 8:30AM-10:30AM
Background/Purpose: Systemic Lupus Erythematosus (SLE) is more common and severe in those of African Ancestry (AA) who, relative to Europeans, are twice as likely to develop lupus nephritis (LN)1. Early nephritis detection can improve outcomes; however, current strategies rely on clinically apparent kidney injury and invasive biopsies, representing a barrier to care in socioeconomically marginalized patients2. Urine sediment epigenetics, which contains invading immune cells, offers a conveniently accessible window into the SLE kidney3, 4.
Methods: This pilot study tested the feasibility of urine epigenetic signature to identify LN in 14 SLE patients in the USA and Nigeria and 2 healthy controls (HC). Consenting participants provided up to 10mL of urine, which was processed fresh (n=4) or treated with 40mM EDTA and 20μl Penicillin/Streptomycin before freezing at -80°C. Samples from Nigeria were shipped on dry ice for processing at the USA site. Urine was centrifuged, and DNA extracted from sediment with the Zymo Research Quick-DNA Urine kit. Methylation of purified DNA was assessed with the Illumina Methylation EPIC BeadChip. Data was analyzed in R using the minfi v1.32.0 software suite. Probe intensities were background corrected and annotated with the IlluminaHumanMethylationEPICanno.ilm10b5.hg38 package. Clustering was performed with principal component analysis (PCA) or multidimensional scaling (MDS) with the top 1000 most variable CpG probes or the 59 control SNP probes on-board the EPIC BeadChip. Intra-sample cell heterogeneity was deconvolved using EpiDISH v2.2.25. Associations between nephritis and differential proportions of each reference cell methylome were tested using a Mann Whitney test.
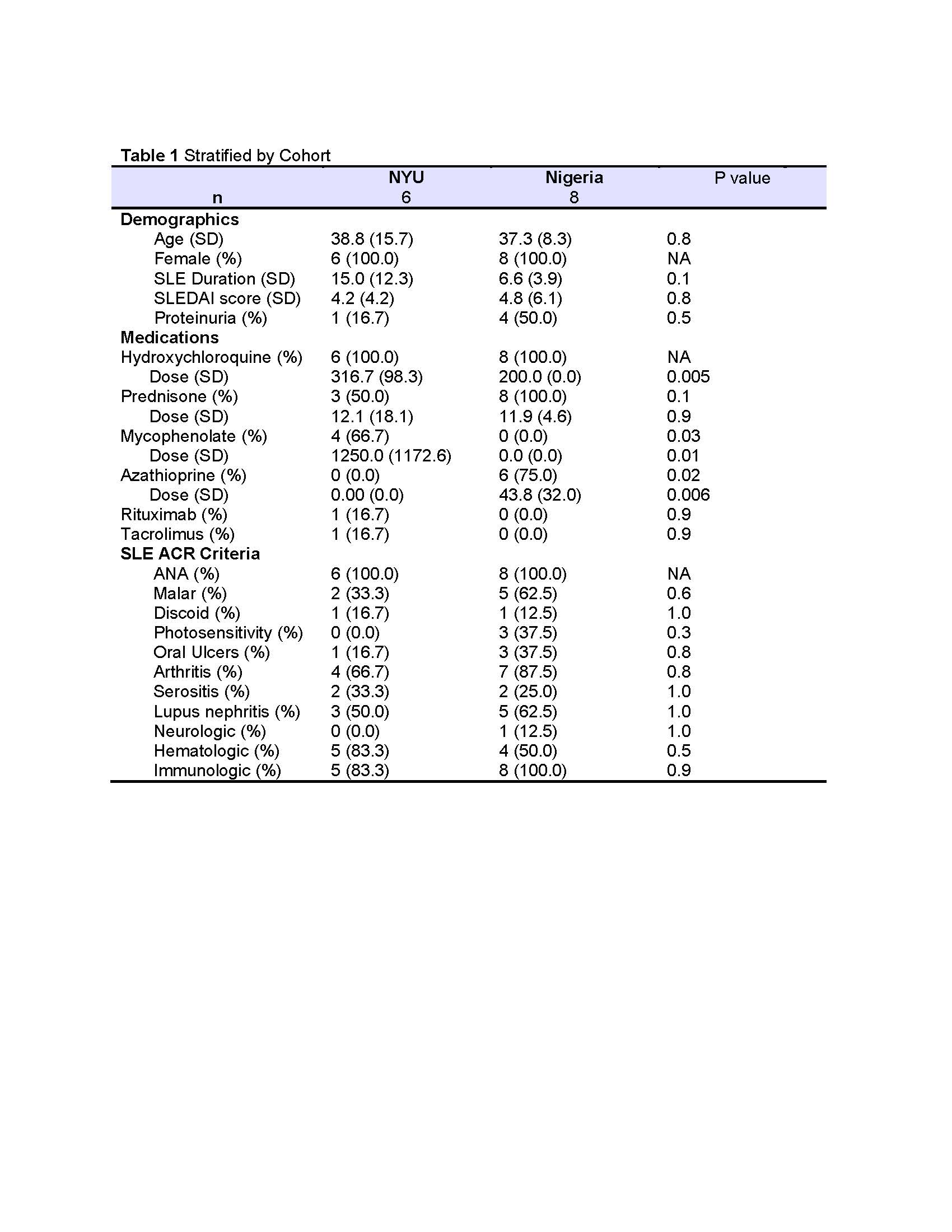
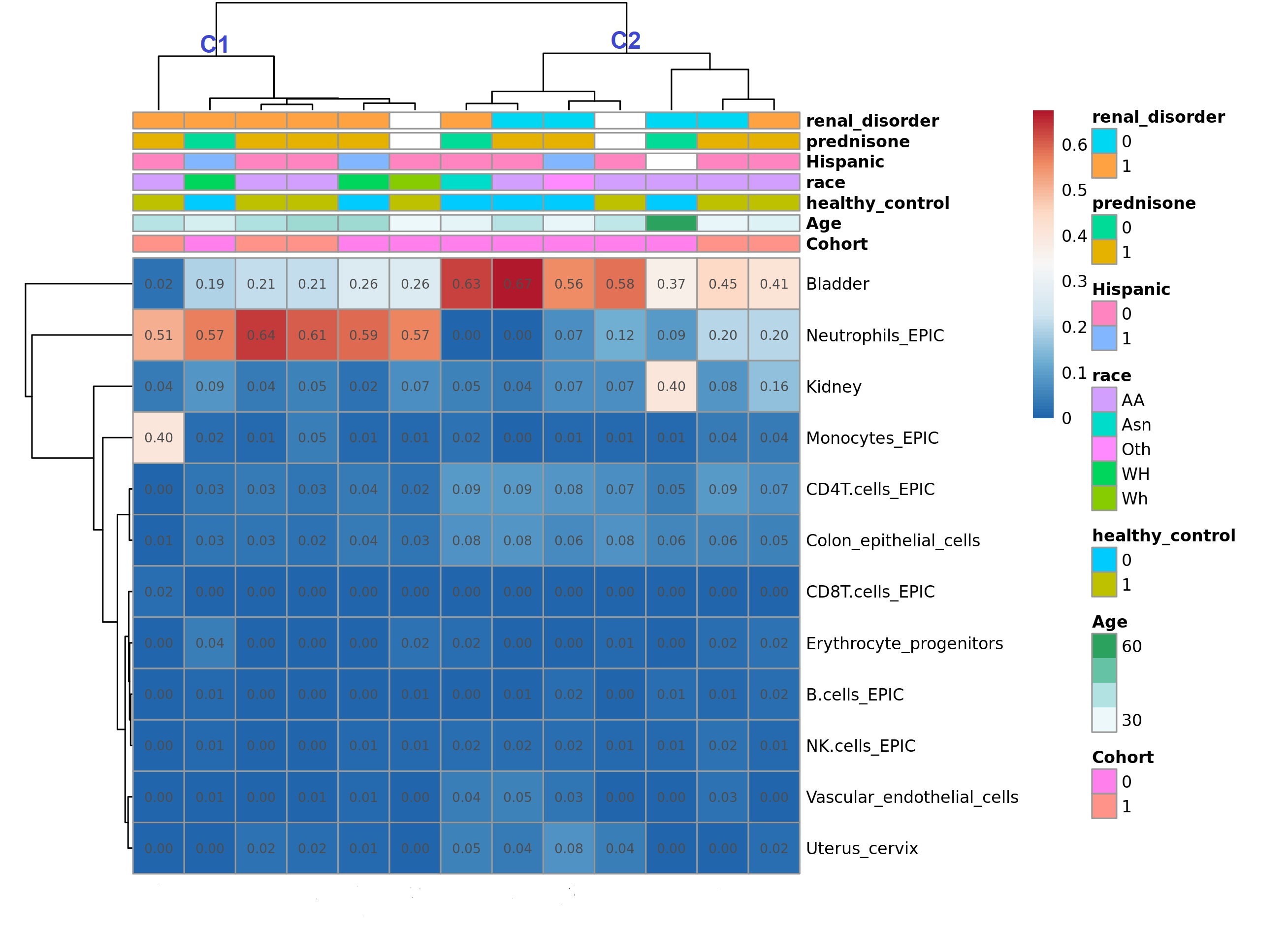
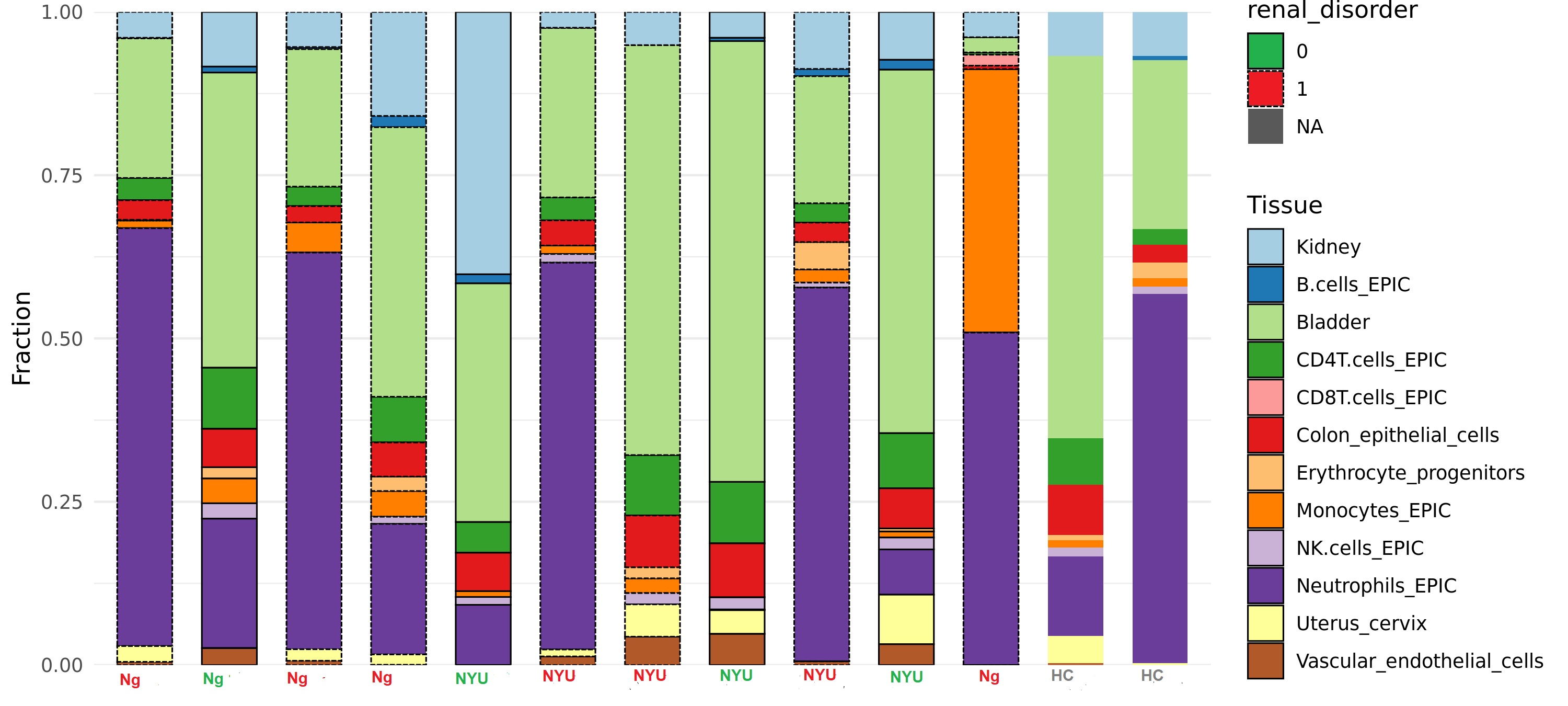
Results: Participant demographics by cohort (USA=6, Nigeria=8) and clinical features are shown in Table 1. In the NYU cohort (and HC), 3 participants were Hispanic, 2 were Asian, and 3 were AA. LN was present in 50% and 62% of the NYU and Nigerian cohorts respectively. At least 100ng of DNA was extracted from 13/16 samples. Fresh, frozen, and shipped-frozen samples produced similar quality DNA; beta distributions and probe intensities were sufficient for CpG calling. PCA analysis of the top 1000 differentially methylated CpG sites revealed clusters by ancestry. Cell type deconvolution revealed that bladder, neutrophils, kidney epithelial, monocytes, CD4 T cells, CD8T Cells, erythrocyte progenitors, B cells, and NK cells were among the most highly represented. A dendrogram of cell type reference fractions produced two major clusters (C1 and C2) which differentiated LN from non-nephritis with 6/7 in C1 and 2/7 in C2 having LN (OR: 12.5, p=0.06; Figure 1). In C2, one LN participant was in complete remission, and the other had predominantly proteinuric disease without active sediment. LN samples were characterized by higher relative fractions of neutrophils and monocytes, and lower fractions of CD4T cells and NK cells compared to non-nephritis samples (Figure 2).
Conclusion: These preliminary data support the feasibility of bulk urine sediment as an epigenetic biomarker of LN, and further analysis looking at pathways and genes within cell proportions associated with LN in larger studies.
To cite this abstract in AMA style:
Blazer A, Ayanlowo O, Liao W, Dey D, Divers J, Ima-Edomwonyin U, Olaosebikan H, Lanata C, Nadkarni G, Buyon J, Izmirly P, Niewold T. EPI-SIGN: Epigenetic SLE Indicators of Glomerular Nephritis [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/epi-sign-epigenetic-sle-indicators-of-glomerular-nephritis/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/epi-sign-epigenetic-sle-indicators-of-glomerular-nephritis/