Session Information
Session Type: Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: To understand the epigenetic patterns of T cells accumulated in rheumatoid arthritis (RA) synovium, we characterized DNA methylation of CD3+ T cells in peripheral blood and synovial tissue in RA and osteoarthritis (OA) patients.
Methods: Genomic DNA of CD3+ T cells was isolated from RA (synovial tissue: n = 8, with 3 matched peripheral blood) and matched OA (5 matched synovial tissue and peripheral blood) patients after separating peripheral blood cells using antibodies and magnetic beads. Methylation was measured using the Illumina Infinium Methylation EPIC Kit chip. Differentially methylated loci (DMLs) were identified using Welch’s t-test and mapped to gene promoter regions to define differentially methylated genes (DMGs). Principal component analysis (PCA) and hierarchical clustering were used to represent the relationship among each group. To compare enriched biological pathways, pathway analysis was applied using Reactome and MSigDB.
Results: PCA results showed that methylation differences in T cells primarily distinguished based on location (blood vs. synovium PCA1 95%) compared with by disease (RA vs OA), mainly with more diversity in synovial T cells (peripheral blood CD3+ cells were clustered within PC3 0.35%) (Figure 1). By comparing DNA methylation differences and DMGs of CD3+ T cells between peripheral blood and synovial tissue, we found 4615 and 832, respectively, for RA but only 164 DMLs and 36 DMGs for OA (q value < 0.05). Therefore, significant blood/synovium methylation differences were mainly identified in RA. By comparing RA and OA to each other, we found 1120 DMLs (p value < 0.05) in 363 DMGs in blood CD3+ T cells and 4104 DMLs (p value < 0.01) in 870 DMGs in synovial CD3+ T cells, respectively. Differentially modified pathways were significantly enriched between RA blood and synovial T cells, especially in genes related to complement, integrin cell surface interactions and the P53 pathway. Only a small number of significantly different pathways were observed between RA and OA peripheral blood. Three significantly enriched pathways between RA and OA in synovial T cells show that the main differences are related to metabolic pathways. Although 870 DMGs were identified between RA and OA in synovial T cells using p value < 0.01, most do not fall into pathways, possibly due to the small sample size. No significant pathways distinguished OA peripheral blood CD3+ T cells compared to OA synovium as only 36 DMGs were identified.
Conclusion: The patterns of DNA methylation in RA show disease and location specific differences in immune pathways including cell contact interactions (compliment and integrins) and survival. The RA joint-specific signatures could be due to selective accumulation of T cell populations, epigenetic modification in situ, or expansion of differentially marked adaptive immune cells. Identifying epigenetic patterns could provide clues to site and event specific targetable T cells in the RA.
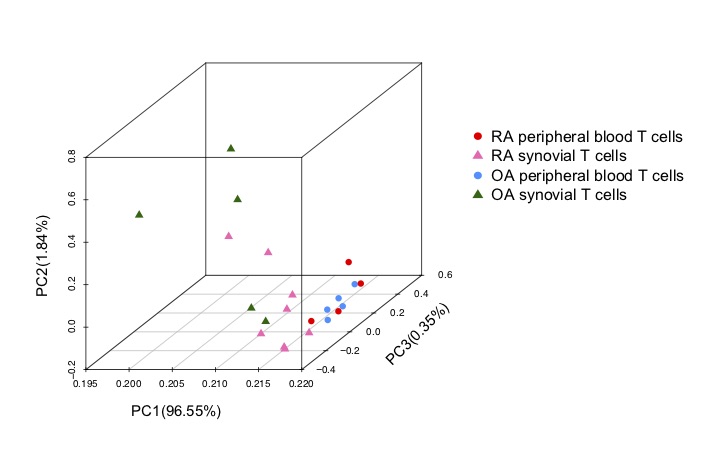 Figure 1. PCA plot shows the segregation of samples are more distinguishable between T cell location (peripheral blood and synovium) than disease. The greatest differences were observed between RA synovial and peripheral blood T cells.
Figure 1. PCA plot shows the segregation of samples are more distinguishable between T cell location (peripheral blood and synovium) than disease. The greatest differences were observed between RA synovial and peripheral blood T cells.
To cite this abstract in AMA style:
Ai R, Firestein G, Boyle D, Wang W. Distinct DNA Methylation Patterns of Rheumatoid Arthritis Peripheral Blood and Synovial Tissue T Cells [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/distinct-dna-methylation-patterns-of-rheumatoid-arthritis-peripheral-blood-and-synovial-tissue-t-cells/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/distinct-dna-methylation-patterns-of-rheumatoid-arthritis-peripheral-blood-and-synovial-tissue-t-cells/
