Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic lupus erythematosus (SLE) is a complex autoimmune disease with heterogeneous clinical manifestations. Understanding the molecular mechanisms of SLE is crucial for developing effective therapeutic strategies.
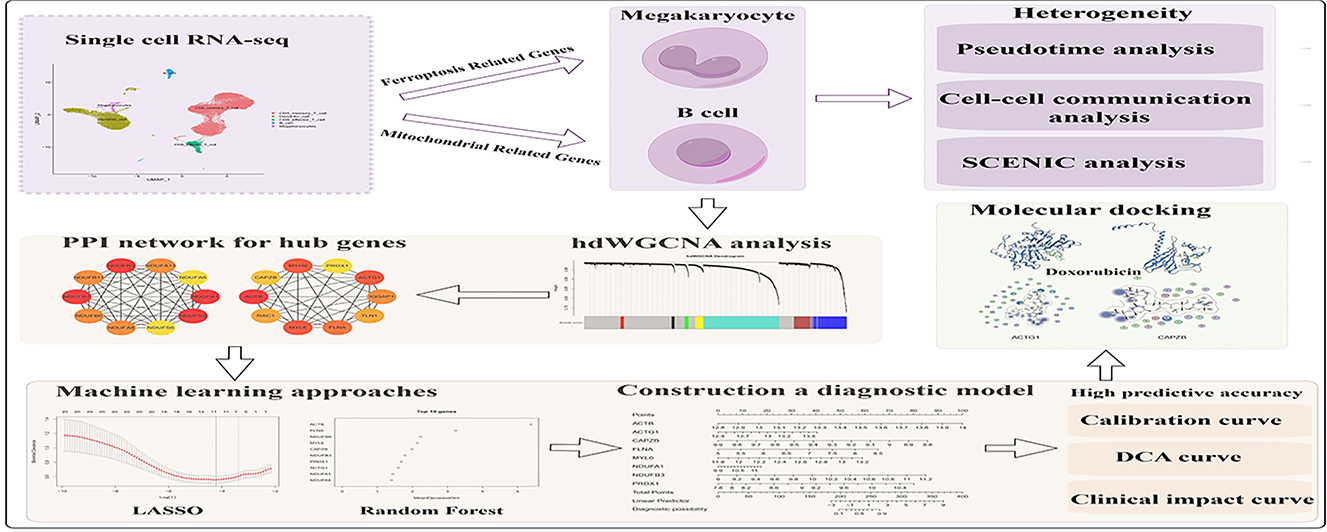
Methods: This study downloaded microarray datasets GSE72326 and GSE50772 from the Gene Expression Omnibus (GEO) database. Single-cell RNA sequencing (scRNA-seq) data from GSE135779 was processed to identify cell types and calculate mitochondrial-related genes (MRGs) and ferroptosis-related genes (FRGs) scores. By employing pseudotime analysis, cell-cell communication analysis, and Single-Cell Regulatory Network Inference and Clustering (SCENIC) analysis, we explored the heterogeneity of cells in SLE. Hub genes were identified using high-dimensional weighted correlation network analysis (hdWGNCA) and machine learning algorithms, leading to the development of a predictive diagnostic model. Additionally, small molecule compounds were screened and docked for potential therapeutic interactions.
Results: We identified 19 clusters and successfully annotated five major cell types. Among these, FRGs scored the highest in Megakaryocytes, while MRGs scored the highest in B cells. Furthermore, we uncovered new cell subtypes and explored their developmental trajectories, intercellular communication patterns, and transcription factors regulating these subpopulations. We also identified eight hub genes and constructed a diagnostic model with high predictive accuracy. Immune infiltration analysis revealed significant correlations between diagnostic biomarkers and various immune cells. Lastly, molecular docking studies suggested Doxorubicin may exert therapeutic effects by affecting these biomarkers.
Conclusion: This study has screened out diagnostic biomarkers for SLE and built the diagnostic model. These findings offer new insights into the pathogenesis of SLE and provide valuable directions for future therapeutic research.
To cite this abstract in AMA style:
Dai Y, Chen Z. Deciphering the Roles of Mitochondrial-Related Genes and Ferroptosis in Systemic Lupus Erythematosus: Integrated Analysis for Diagnostic Biomarker Identification and Therapeutic Insights [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/deciphering-the-roles-of-mitochondrial-related-genes-and-ferroptosis-in-systemic-lupus-erythematosus-integrated-analysis-for-diagnostic-biomarker-identification-and-therapeutic-insights/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/deciphering-the-roles-of-mitochondrial-related-genes-and-ferroptosis-in-systemic-lupus-erythematosus-integrated-analysis-for-diagnostic-biomarker-identification-and-therapeutic-insights/