Session Information
Date: Tuesday, November 9, 2021
Title: Osteoarthritis & Joint Biology – Basic Science Poster (1468–1479)
Session Type: Poster Session D
Session Time: 8:30AM-10:30AM
Background/Purpose: Strong links between epigenetic changes, particularly alterations in DNA methylation, have been linked with the onset and progression of knee osteoarthritis (OA); however, the environmental factors driving epigenetic changes within articular tissues have not yet been fully elucidated. We have previously demonstrated that the microbial DNA signatures within human cartilage and shown changes in microbial signatures correlate with primary knee OA progression. In this study, we hypothesized that alterations in microbial DNA may induce epigenetic changes within articular cartilage. To test this, we exposed chondrocyte cell lines in vitro to amplified microbial DNA from human OA patients and controls.
Methods: Matched discarded cartilage samples from macroscopically eroded (n=4) and intact (n=4) OA specimens were obtained from patients undergoing total knee arthroplasty. Control cartilage samples were obtained from cadaveric control patients. Microbial DNA was isolated using an anti-MBD2 magnetic bead approach and microbial DNA subjected to whole-genome amplification. Twenty micrograms of amplified microbial DNA was added Tc28a2 cells in culture. After a 3-day incubation period, DNA was extracted, treated with sodium bisulfite, and loaded onto Illumina Infinium Methylation EPIC chips .DNA methylation beta values were extracted from raw chip imaging and processed in R using the ChAMP package. Genes associated with differentially methylated CpG sites were evaluated ontologically using the Ingenuity Pathway Analysis (IPA) software package.
Results: Microbial DNA treatment induced substantial changes in DNA methylation levels. Comparing eroded-OA microbial DNA-treated cells with control microbial DNA-treated cells, we found 1638 differentially methylated CpG sites (DMPs), 1637 hypomethylated in the eroded group, corresponding to 1109 unique genes. No DNA methylation differences were induced when comparing non-OA with intact-OA microbial DNA-treated cells. Comparing eroded-OA to intact-OA, we identified 1275 DMPs, 1274 hypomethylated in the eroded group, corresponding to 881 unique genes. Ontology analysis of differentially methylated genes associated with eroded-OA vs. control microbial DNA treatment revealed numerous OA-related pathways and upstream regulators (Figure 1). These include axonal guidance signaling, Rho family GTPases, ERK/MAPK signaling, stem cell pluripotency, and ErbB signaling, among others. Upstream analysis included enrichment in ERG, miR-137 (which regulates ADAMTS5), miR-21 (which targets Gdf5), miR-16 (which targets Smad3), among others.
Conclusion: Herein, we demonstrate that exposure of human chondrocytes in vitro to microbial DNA is sufficient to induce widespread epigenetic changes. OA-related epigenetic modifications are identified when cells are treated with microbial DNA extracted from eroded-OA human cartilage samples. These data represent a novel potential mechanism of microbiome-epigenome interaction in the development of OA. Future work should focus on testing whether these findings are replicable in vivo and explore the mechanism(s) through which this epigenetic alteration is induced.
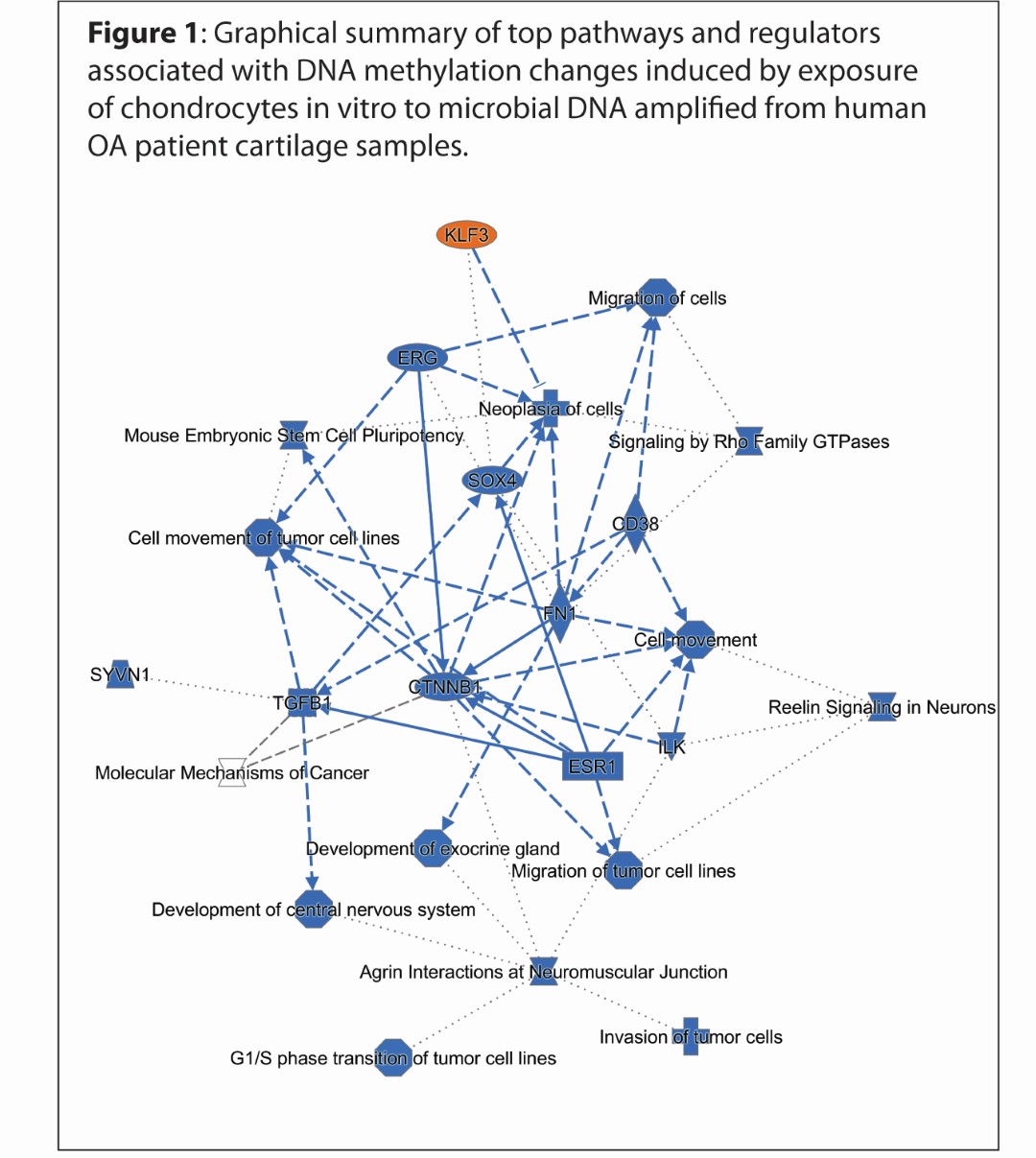 Figure 1: Graphical summary of top pathways and regulators associated with DNA methylation changes induced by exposure of chondrocytes in vitro to microbial DNA amplified from human OA patient cartilage samples.
Figure 1: Graphical summary of top pathways and regulators associated with DNA methylation changes induced by exposure of chondrocytes in vitro to microbial DNA amplified from human OA patient cartilage samples.
To cite this abstract in AMA style:
Izda V, Dunn C, Sturdy C, Martin J, Velasco C, Jacob P, Jeffries M. Cartilage Epigenetic Changes Induced by Microbial DNA Amplified from Human OA Samples [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/cartilage-epigenetic-changes-induced-by-microbial-dna-amplified-from-human-oa-samples/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/cartilage-epigenetic-changes-induced-by-microbial-dna-amplified-from-human-oa-samples/
