Session Information
Session Type: Abstract Session
Session Time: 9:00AM-9:15AM
Background/Purpose: It is widely accepted that activation of specific CD4 T cells through their TCRs by self-antigen (Ag) is necessary for rheumatoid arthritis (RA) onset. Yet, the transcriptional profiles and TCR identities of the disease-causing pathogenic CD4 T cells remain unknown. SKG is an ideal platform to study questions relevant to RA since this model not only captures many of the important features of human RA, but also recapitulates the paradoxical ability of CD4 T cells to differentiate into pathogenic effector cells despite their impaired response to TCR engagement. We combined SKG mice with a fluorescent reporter tethered to the regulatory region of Nr4a1 (Nur77—which is rapidly and selectively upregulated in response to Ag, but not inflammatory stimuli) to read out the relative strength of TCR signaling (termed by us SKGNur mice). Using these mice we previously identified a CD4 T cell subpopulation that is greatly enriched for their arthritogenic potential (in the GFPhi fraction). In this study, we examine the T cell repertoire and signaling networks invoked by endogenous Ag encounter in pre-arthritogenic T cells to study the evolution of autoimmune arthritis.
Methods: We used bulk and paired single-cell RNA and TCR-sequencing (scRNAseq and TCRseq) to simultaneously profile the transcriptome and TCR repertoire of self-reactive pathogenic T cells before arthritis onset in SKGNur mice. Analysis was performed on sorted naïve SKG and BALB/c (WT) CD4 T cells based on their Nur77-eGFP levels (GFPhi and GFPlo) using Q2 Solutions and the 5’ 10X platform respectively. After filtering of the latter, there were 99,074 cells with a mean of 12,384 cells per condition. We examined our datasets for differentially expressed genes (DEG) and TCR variable genes enriched in the arthritogenic SKGNur GFPhi T cells. Flow cytometry was used to validate genes that were found to be up or down-regulated in arthritogenic T cells.
Results: In our bulk RNAseq dataset >990 DEGs were identified and clustered into 6 gene modules that capture the transcriptional differences between the SKGNur and WTNur GFPhi and GFPlo subgroups. Gene set enrichment analysis revealed a T cell activation signature in the naïve SKGNur GFPhi CD4 T cells. Our scRNAseq dataset revealed significant heterogeneity within the unperturbed naïve CD4 T cell compartment with 8 distinct clusters further refining our bulk RNAseq results. We found that T cells expressing the highest levels of Nr4a1 (red cluster in Fig 1) upregulate TCR signaling gene modules, which are enhanced in SKG mice despite their hypomorphic mutation in ZAP70, a tyrosine kinase critical for proximal TCR signaling. In simultaneous investigation of their TCR sequences, we uncovered a previously unknown biased TCR Vb usage in the SKG repertoire (Fig 2, DNS). The Vb genes whose expression is uniquely enriched in the SKGNur GFPhi subset are well known to recognize endogenous mouse mammary tumor virus superantigen. We detect still further enrichment of these Vb’s in the SKG arthritic joint (Fig 2, DNS).
Conclusion: Our results suggest both transcriptional activation and repertoire selection driven by endogenous viral superantigen(s) act as inter-related mechanisms that prime arthritogenic SKG T cells prior to disease onset.
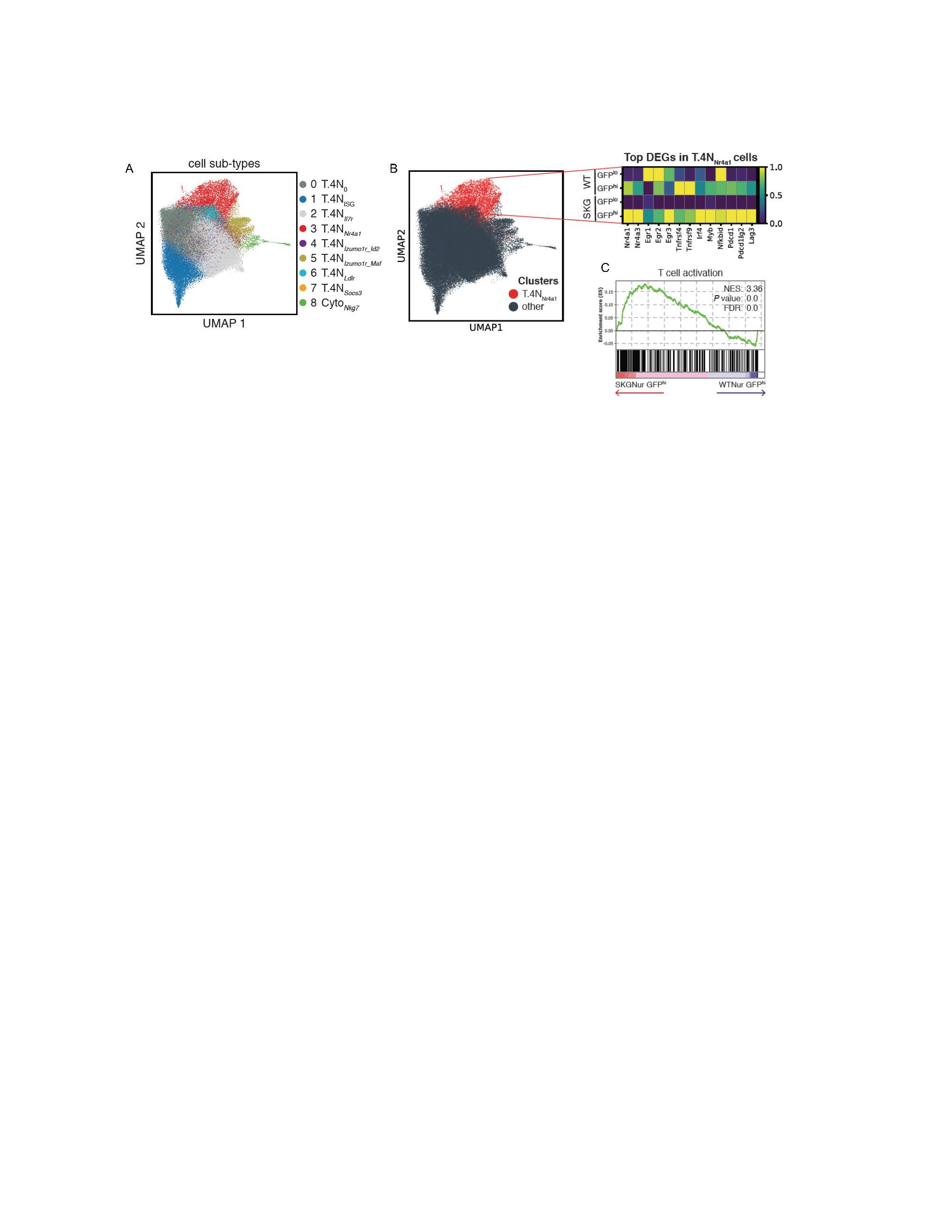 Figure 1. Arthritogenic SKG T cells upregulate a transcriptional TCR signaling program. (A) UMAP of 99,074 naïve T cells derived from 8 samples (2 replicates from each WT and SKG GFPlo and GFPhi subset); cells colored according to annotated leiden clusters. (B) Heatmap shows average expression of top DEGs (abs value(log2(FC)) > 2.8, adj P value < 0.05) of cells in T.4N_Nr4a1 (Nr4a1 high) cluster vs other cells by subgroup normalized by standard scale by column. (C) Enrichment plot of T cell activation pathway from GSEA analysis for ranked genes in T.4N_Nr4a1 cells from SKG GFPhi v. WT GFPhi differential expression analysis. FDR, false discovery rate. NES, normalized enrichment score.
Figure 1. Arthritogenic SKG T cells upregulate a transcriptional TCR signaling program. (A) UMAP of 99,074 naïve T cells derived from 8 samples (2 replicates from each WT and SKG GFPlo and GFPhi subset); cells colored according to annotated leiden clusters. (B) Heatmap shows average expression of top DEGs (abs value(log2(FC)) > 2.8, adj P value < 0.05) of cells in T.4N_Nr4a1 (Nr4a1 high) cluster vs other cells by subgroup normalized by standard scale by column. (C) Enrichment plot of T cell activation pathway from GSEA analysis for ranked genes in T.4N_Nr4a1 cells from SKG GFPhi v. WT GFPhi differential expression analysis. FDR, false discovery rate. NES, normalized enrichment score.
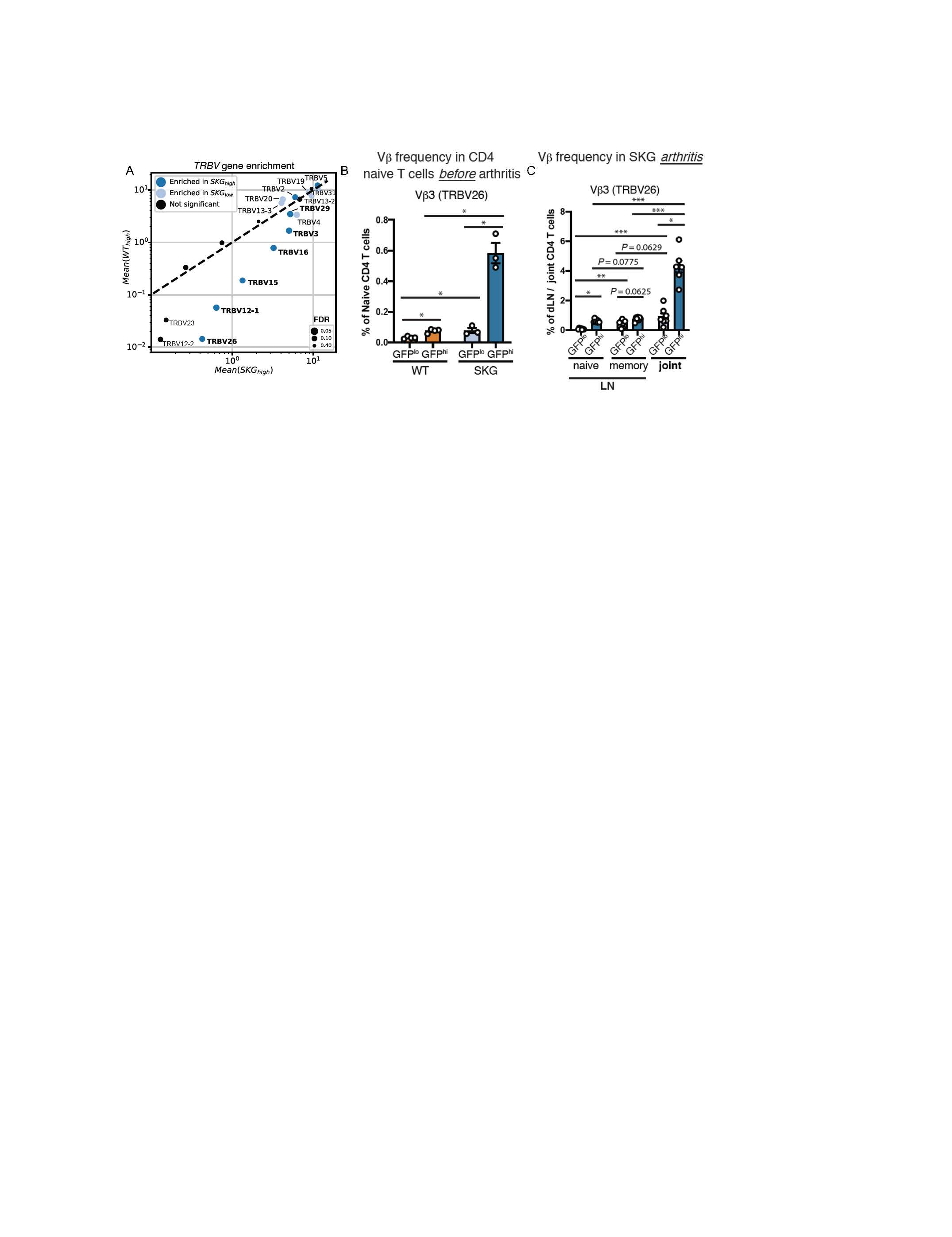 Figure 2. SKG T cells harbor a biased TCR Vb repertoire likely pruned by endogenous superantigen(s). (A) Scatterplot of mean frequency of cells expressing each TCR variable beta gene (TRBV) gene for SKGNur vs WTNur GFPhi. Dots for each TRBV gene are sized according to FDR from one-sided paired t-test comparing frequency in SKG GFPhi vs SKG GFPlo. Dots colored as either significantly enriched (FDR < 0.1) in SKG GFPhi (dark blue), SKG GFPlo (light blue), or not significantly enriched in either subgroup (black). Labels for TRBV genes significantly enriched in SKG GFPhi and more highly expressed in SKG GFPhi vs WT GFPhi samples are bolded. (B-C) Mean frequency (± SEM) of indicated CD4 T cell subset with TCR Vb3 protein usage determined by flow cytometry in GFPlo and GFPhi T cells from lymph nodes (LN) prior to arthritis induction (B) or LN and joints 2.5 weeks after arthritis (C), repeated > 2 times.
Figure 2. SKG T cells harbor a biased TCR Vb repertoire likely pruned by endogenous superantigen(s). (A) Scatterplot of mean frequency of cells expressing each TCR variable beta gene (TRBV) gene for SKGNur vs WTNur GFPhi. Dots for each TRBV gene are sized according to FDR from one-sided paired t-test comparing frequency in SKG GFPhi vs SKG GFPlo. Dots colored as either significantly enriched (FDR < 0.1) in SKG GFPhi (dark blue), SKG GFPlo (light blue), or not significantly enriched in either subgroup (black). Labels for TRBV genes significantly enriched in SKG GFPhi and more highly expressed in SKG GFPhi vs WT GFPhi samples are bolded. (B-C) Mean frequency (± SEM) of indicated CD4 T cell subset with TCR Vb3 protein usage determined by flow cytometry in GFPlo and GFPhi T cells from lymph nodes (LN) prior to arthritis induction (B) or LN and joints 2.5 weeks after arthritis (C), repeated > 2 times.
To cite this abstract in AMA style:
ASHOURI J, McCarthy E, Yu S, Perlmutter N, Lin C, Yu C, Weiss A. Arthritogenic T Cells Harbor a Transcriptional Program of T Cell Activation and a Repertoire Pruned by Endogenous Superantigen [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/arthritogenic-t-cells-harbor-a-transcriptional-program-of-t-cell-activation-and-a-repertoire-pruned-by-endogenous-superantigen/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/arthritogenic-t-cells-harbor-a-transcriptional-program-of-t-cell-activation-and-a-repertoire-pruned-by-endogenous-superantigen/
