Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Gout affects >12 million US adults and is associated with premature all-cause and cardiovascular (CV) mortality which has failed to improve over recent decades, unlike in the general population.1,2 Comorbidities and treatment inequity contribute, but underlying gout disease-specific mechanisms may also play a role, since the elevated mortality persists with adjustment for cardiometabolic comorbidities, kidney function, BMI, alcohol, all atherosclerotic cardiovascular (ASCVD) risk factors, and serum urate (SU) levels.1 Since detailed characterisation of the plasma proteome can illuminate mechanisms and risk stratification of premature mortality in gout, we leveraged large-scale proteomics data for a nationwide prospective cohort.
Methods: We studied 2,013 UK Biobank participants with diagnosis of gout and proteomic profiling available (mean age 60 years, 83% males). Proteomic measures (n=2923, via Olink Explore 3072) were quantified from baseline blood samples via Proximity Extension Assay technology. After division into training (80%) and testing (20%) sets, least absolute shrinkage and selection operator (LASSO) regression with 10-fold cross validation was used to identify most robust prognostic proteins and construct a proteomic risk score (signature) for mortality in the training set. Multivariable Cox models were compared between clinical factors (age, sex, race, alcohol, other ASCVD risk factors, BMI, cardiometabolic comorbidities, kidney function, diuretic medications, and SU levels), with and without the proteomic signature, using area under the receiver operating characteristic curve (AUC). Two-sided DeLong test was used to compare AUCs of each model in training and testing sets.
Results: We documented 469 deaths over 25,751 person-years (PY) of follow-up (18.2 per 1000 PY) in the gout cohort. In the training set (386 deaths), clinical factors alone discriminated mortality with an AUC of 0.717 (95% CI: 0.698, 0.737). From 790 individually-associated proteins (PFDR < 0.05), LASSO regression generated a 76-protein signature of mortality risk; each unit increase in risk score was associated with 2.5-fold greater mortality risk (Table 1). Highest-weighted markers in the signature included NT-proBNP, growth-differentiation factor 15 (biomarker of major organ injury, cardiac and cancer risk, and predictive factor for CV disease and mortality), and TNF-R superfamily member 10A (TNFRSF10A) (Table 2). Excess TNFRSF10A transduces apoptosis and marked release of multiple inflammatory cytokines (e.g., IL-8, TNF, and CCL20). Addition of the proteomic signature to the clinical model significantly improved discrimination over the clinical model alone (P < 0.001), with AUC of 0.856 (0.844, 0.868) (Figure). Proteomic signature also performed well in the testing set (83 deaths), with AUC 0.672 (0.639, 0.705) for the clinical model alone and 0.812 (0.785, 0.840) for the combined.
Conclusion: Addition of a proteomic signature improves ability to predict risk of all-cause mortality among individuals with gout. The component proteomic biomarkers may allow for risk prediction and stratification, and identification of potential novel therapeutic targets. 1PMID 38191784; 2PMID 28122760
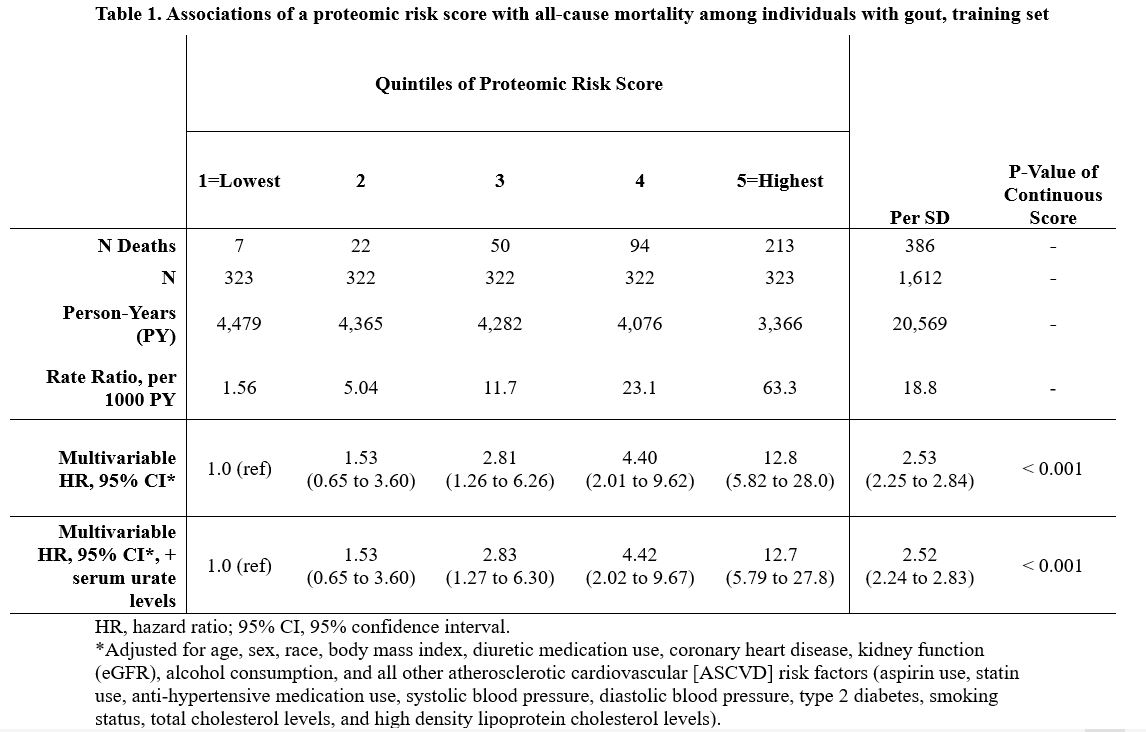 Table 1. Associations of a proteomic risk score with all-cause mortality among individuals with gout, training set
Table 1. Associations of a proteomic risk score with all-cause mortality among individuals with gout, training set
.jpg) Table 2. Proteomic biomarkers associated with all-cause mortality among individuals with gout, five highest-weighted associations in the proteomic risk score
Table 2. Proteomic biomarkers associated with all-cause mortality among individuals with gout, five highest-weighted associations in the proteomic risk score
.jpg) Figure: Receiver operating characteristic curves for discrimination of all-cause mortality with clinical factors and a proteomic signature. AUC= area under the receiver operating characteristic curve.
Figure: Receiver operating characteristic curves for discrimination of all-cause mortality with clinical factors and a proteomic signature. AUC= area under the receiver operating characteristic curve.
Clinical factors include age, sex, race, body mass index, alcohol consumption, diuretic medication use, coronary heart disease, kidney function (eGFR), atherosclerotic cardiovascular (ASCVD) risk factors (aspirin use, statin use, anti-hypertensive medication use, systolic blood pressure, diastolic blood pressure, type 2 diabetes, smoking status, total cholesterol levels, high density lipoprotein cholesterol levels), and serum urate levels.
To cite this abstract in AMA style:
McCormick N, Rai S, Yokose C, Merriman T, Terkeltaub R, Choi H. A Proteomic Signature Containing TNF Receptor Superfamily Member 10A (TNFRSF10A) and Growth/Differentiation Factor 15 (GDF-15) Improves Prediction of All-Cause Mortality Among Individuals with Gout, Beyond Atherosclerotic Cardiovascular and Other Clinical Risk Factors [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/a-proteomic-signature-containing-tnf-receptor-superfamily-member-10a-tnfrsf10a-and-growth-differentiation-factor-15-gdf-15-improves-prediction-of-all-cause-mortality-among-individuals-with-go/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-proteomic-signature-containing-tnf-receptor-superfamily-member-10a-tnfrsf10a-and-growth-differentiation-factor-15-gdf-15-improves-prediction-of-all-cause-mortality-among-individuals-with-go/
