Session Information
Date: Monday, October 27, 2025
Title: (0978–1006) T Cell Biology & Targets in Autoimmune & Inflammatory Disease Poster
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Cutaneous lupus erythematosus (CLE) is a frequent manifestation of systemic lupus erythematosus (SLE) and remains an important contributor to morbidity in lupus patients. Despite its clinical significance, the underlying mechanisms of CLE and its pathophysiological links to SLE are poorly understood. Importantly, new data support a critical difference between cutaneous and peripheral immune subsets in the same patient. This study aimed to elucidate the shifts in T cell and natural killer cell subsets from blood to lupus-affected skin.
Methods: We performed single-cell RNA sequencing of non-lesional and lesional skin biopsies and paired PBMCs from 17 patients with active CLE/SLE and non-lesional skin and PBMCs from 5 healthy controls via 10x Genomics pipelines. Cellular clustering and annotation were conducted using the Seurat R package by matching cluster-defining marker genes with canonical subtype signatures. To uncover key upstream regulators, we applied Ingenuity Pathway Analysis (IPA) to differentially expressed genes (DEGs). Cytokines and transcription factors with an activation z-score ≥2 or ≤–2 were considered significant regulators. Pseudotime trajectory was constructed using the R package Monocle.
Results: Visualization using uniform manifold approximation and projection (UMAP) revealed 30 distinct cell clusters (Figure 1). We identified transcription factor NONO and proinflammatory cytokines including type I interferons (IFNs), IL-1, IL-27, TNF and prolactin as primary upstream regulators of transcripts enriched in regulatory T cells (Tregs), interferon-educated T (IFN-T) cells, and natural killer (NK) cells in lupus skin. These effects were observed when compared to both paired peripheral blood and healthy control skin (Figure 2), with the signaling being most pronounced in lesional as compared to non-lesional lupus skin. In contrast, the signaling of TGF-β, IL-4, MLXIPL, MYC, MYCN, SPEN, ETV3, and ETV6 was inhibited in Tregs in lupus skin compared to healthy controls. Furthermore, NONO and type I IFNs were important upstream regulators of gene expression in CD69lo NK cells, while IL-1 and TGF-β were stronger regulators of gene expression in CD69hi NK cells in skin. Trajectory analysis showed that circulating NK cells, IFN-T cells and Tregs upregulate CD69 whereas Tregs additionally acquire the expression of CTLA-4, GATA-3, heat shock protein, PD-1 and BATF as they infiltrate the skin.
Conclusion: Our findings highlight the role of the skin in shaping an inflammatory profile in lupus Treg, IFN-T, and NK cell subsets. Treg development in lupus skin appears to shift from TGF-β- to type I IFN–mediated signaling. While NK cells, IFN-T cells and Tregs acquire activated phenotypes, Tregs additionally develop potentially exhausted phenotypes in SLE. NONO and type I IFNs contribute to the early programming of NK cells, while IL-1 drives their activation. These insights provide a deeper understanding of T cell–driven inflammation in CLE and may inform future therapeutic strategies.
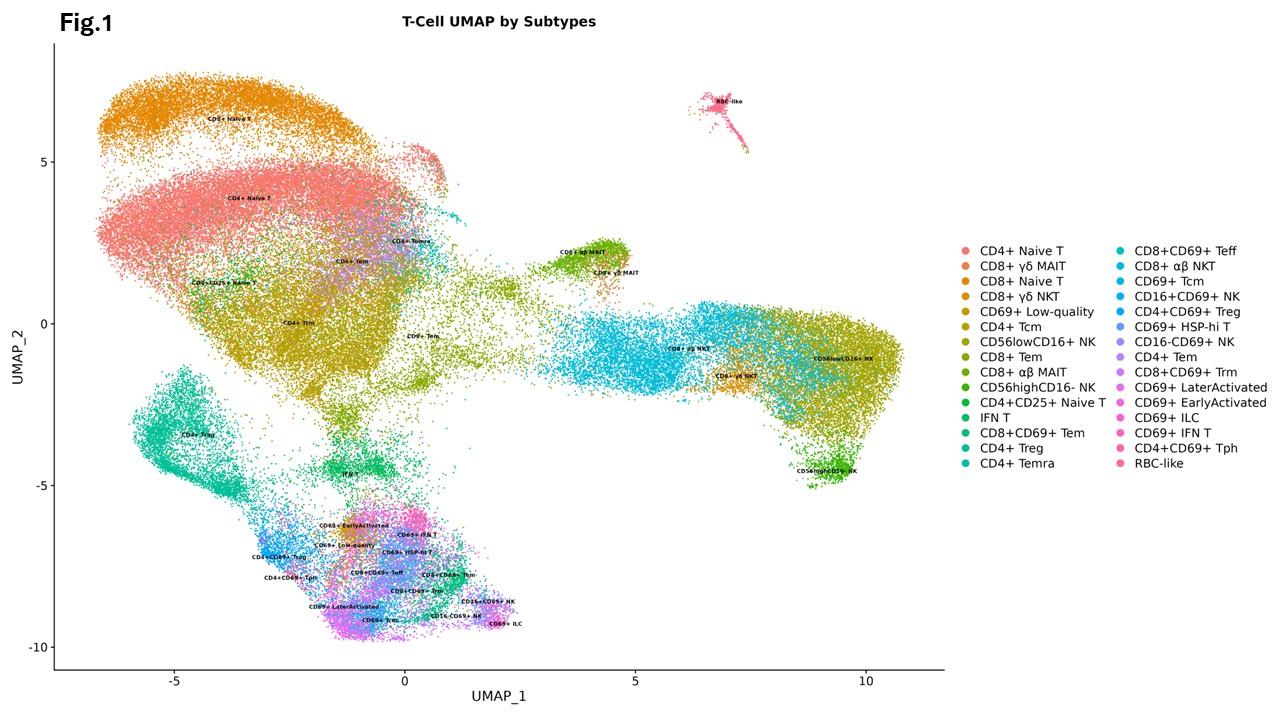 UMAP plot of 92,405 cells colored by cluster
UMAP plot of 92,405 cells colored by cluster
.jpg) Dot plot of the top upstream regulators enriched among DEGs in Skin versus peripheral blood SLE Tregs
Dot plot of the top upstream regulators enriched among DEGs in Skin versus peripheral blood SLE Tregs
To cite this abstract in AMA style:
Kato H, Zhang L, Gharaee-Kermani M, Hurst A, Bogle R, Tsoi A, Gudjonsson J, Kahlenberg J. Lupus Skin Shapes a Distinct Inflammatory Milieu that Drives the Skewing of Treg and inflammatory T cells [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/lupus-skin-shapes-a-distinct-inflammatory-milieu-that-drives-the-skewing-of-treg-and-inflammatory-t-cells/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/lupus-skin-shapes-a-distinct-inflammatory-milieu-that-drives-the-skewing-of-treg-and-inflammatory-t-cells/
