Session Information
Date: Monday, October 27, 2025
Title: (0955–0977) Systemic Sclerosis & Related Disorders – Basic Science Poster I
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Regression of skin fibrosis is a feature of the natural history of dcSSc. The molecular mechanisms underlying this resolution remain unclear. This study aims to establish a molecular fingerprint characterizing regression of skin fibrosis in SSc patients.
Methods: All patients fulfilled the 2013 ACR/EULAR classification criteria for SSc. Regressors were defined by a 5-point decrease in the modified Rodnan’s Skin Score (mRSS), and progressors by a 5-point increase at annual follow-up. Serum proteomics using the Olink High Throughput Explore panel (5,400 proteins) was performed on 44 SSc patients (25 regressors, 19 stable/progressors=non-regressors). Differentially abundant proteins (DAPs) were identified using the limma package. For single-cell RNA sequencing (scRNA-seq), peripheral blood mononuclear cells (PBMCs) were isolated from 15 SSc patients (5 each: regressor, progressor, stable) and 5 healthy controls. Single-cell libraries were prepared, and raw reads were aligned to the human genome using Cellranger 7.0. Transcriptomic data were analyzed with Seurat (v5.1.0), and differential gene expression analysis (DGEA) was performed using the MAST package, followed by Gene Set Enrichment Analysis (GSEA).
Results: Olink proteomics analysis identified 124 DAPs between skin regressors and skin non-regressors. Pathway enrichment analysis (PEA) on the DAPs revealed that MDK, THY1, JAM3, XG and VEGFB relate to cellular extravasation processes and SMOC2, RAMP2, CD160, VEGFB, HSPG2, PGF, EPHA2 to angiogenesis as top processes (Fig. 1). Seventy-three percent of these proteins are linked to monocyte extravasation. PBMC scRNA-seq revealed 5 distinct clusters of monocytes (Fig 2A). Compositional analysis revealed a significant increase of clusters 0, 1 and 3 in the regressors compared to progressors (Fig 2B). To find whether a specific subset is involved in monocyte-vascular interactions, the expression of extravasation markers ITGAM, SELL, ICAM1, and S100A8/9 was used. These markers localized to clusters 0, 1 and 3 (Fig 2C). Additionally, chemotaxis marker CCR1 had an increased expression in clusters 0, 1, and 3, whereas CCR5 was unique to cluster 1 (Fig 2D). Further characterization of cluster 1 using GSEA revealed a suppression in the gene sets ‘vasculature development’ (including extravasation-related genes: ITGA5, MMP14, MMP2, MMP19, CCL2, SOCS3, HMOX1 and PPARG) and ‘cell adhesion’ (including extravasation-related genes: ITGA5, MMP14, MMP2, CCL2, SOCS3, SIGLEC1 and MERTK). The generated gene module scores reflect a consistent decrease in regressors compared to other patient groups (Fig 3A-C). Additionally, both PEA and GSEA on cluster 1 had a decreased expression of IFN-I genes in the regressors, which is linked to a decrease in vascular permeability (Fig 3D).
Conclusion: Our multi-omics approach identified a set of proteins linked to monocyte extravasation and a subcluster of monocytes characterized by extravasation markers. This subcluster also displayed a decreased IFN-I activity. Our data points to a reduction in monocyte extravasation to the tissue associated to the regression of skin fibrosis in SSc.
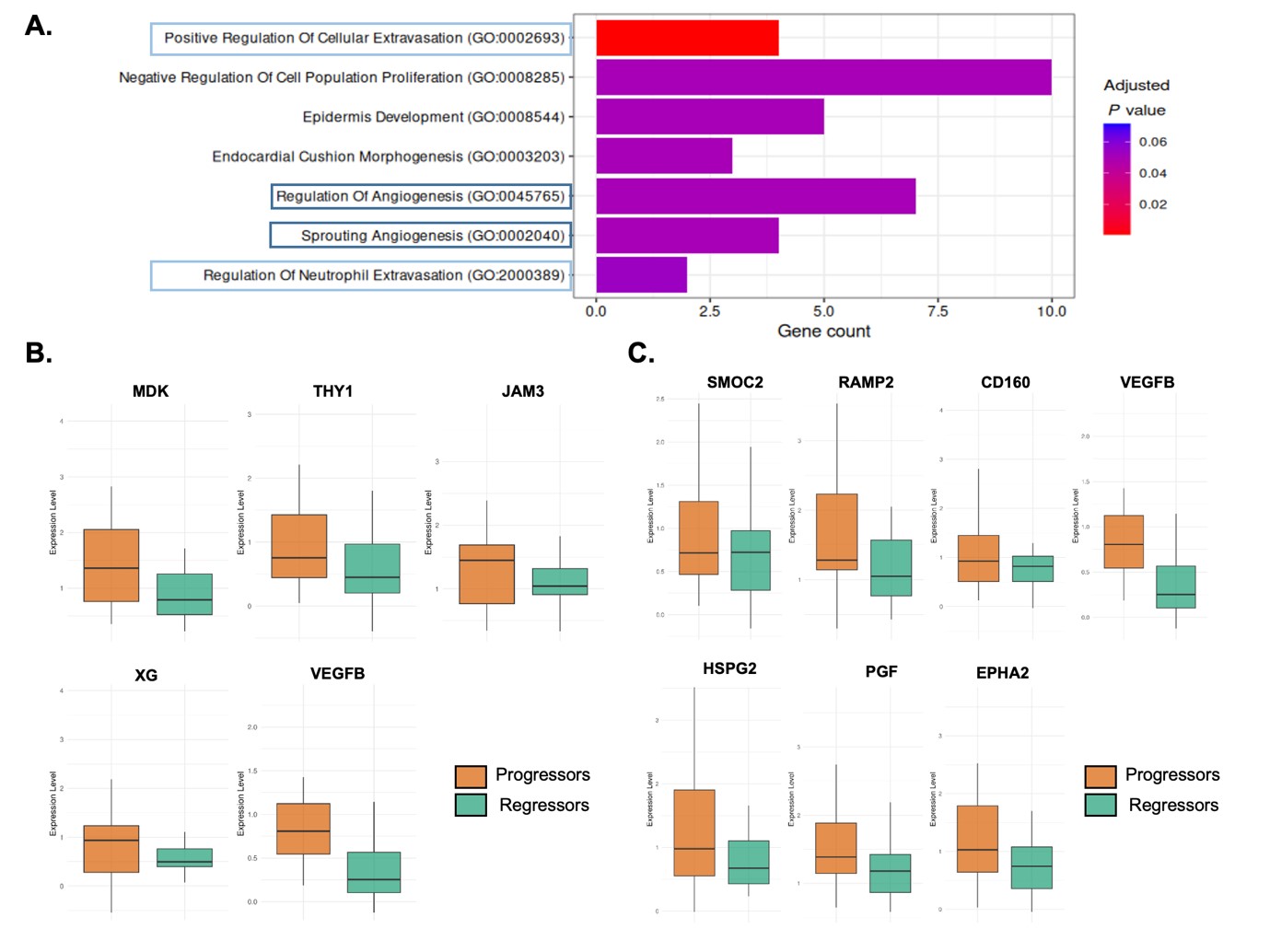 Figure 1. Serum proteomics reveals that differentially abundant proteins (DAPs) are linked to cellular extravasation and angiogenesis and downregulated in regressors. A) Enrichment analysis using GO Biological Processes of the 124 DAPS detected (Benjamini-Hochberg adjusted FDR < 0.3) shows that DAPs are linked to processes including ‘Positive Regulation of Cellular Extravasation’ and ‘Regulation of Angiogenesis’. B) The DAPs linked to the cellular extravasation process all demonstrate a decreased expression in the regressors compared to the progressors. C) DAPs linked to the angiogenesis process show a decreased expression in the regressors compared to the progressors.
Figure 1. Serum proteomics reveals that differentially abundant proteins (DAPs) are linked to cellular extravasation and angiogenesis and downregulated in regressors. A) Enrichment analysis using GO Biological Processes of the 124 DAPS detected (Benjamini-Hochberg adjusted FDR < 0.3) shows that DAPs are linked to processes including ‘Positive Regulation of Cellular Extravasation’ and ‘Regulation of Angiogenesis’. B) The DAPs linked to the cellular extravasation process all demonstrate a decreased expression in the regressors compared to the progressors. C) DAPs linked to the angiogenesis process show a decreased expression in the regressors compared to the progressors.
.jpg) Figure 2. Monocytes in scRNA-seq display a signature related to monocyte extravasation that can be linked to cluster 1. A) UMAP of all monocytes in the PBMC scRNA-seq data revealed 5 distinct monocyte subpopulations. B) Compositional analysis shows that cluster 1 is significantly more abundant in the regressors compared to progressors (FDR = 0.11). C) Well-established markers related to cellular extravasation (ITGAM, SELL, CX3CR1, ICAM1, S100A8, S100A9) showed an increased transcript expression in cluster 1. D) CCR1, which mediates monocyte arrest showed increased expression in clusters 0, 1, and 3. CCR5, which is involved in monocyte spreading, shows a specific expression in only cluster 1.
Figure 2. Monocytes in scRNA-seq display a signature related to monocyte extravasation that can be linked to cluster 1. A) UMAP of all monocytes in the PBMC scRNA-seq data revealed 5 distinct monocyte subpopulations. B) Compositional analysis shows that cluster 1 is significantly more abundant in the regressors compared to progressors (FDR = 0.11). C) Well-established markers related to cellular extravasation (ITGAM, SELL, CX3CR1, ICAM1, S100A8, S100A9) showed an increased transcript expression in cluster 1. D) CCR1, which mediates monocyte arrest showed increased expression in clusters 0, 1, and 3. CCR5, which is involved in monocyte spreading, shows a specific expression in only cluster 1.
.jpg) Figure 3. Characterization of Cluster 1 reveals a subpopulation of monocytes primed for extravasation. A) GSEA showed a suppression of gene sets ‘regulation of endothelial cell proliferation’ ad ‘vasculature development’ in the regressors. Similarly, gene sets ‘regulation of type I interferon production’ and ‘type I interferon production’ were also suppressed in the regressors. B) The gene module score for the vasculature development gene set was calculated using the aggregated expression of the genes in this gene set. The gene module score for ‘Vasculature Development’-associated genes and ‘Endothelial Cell Proliferation’-related genes D) were both found to be suppressed in the regressors compared to other patient groups. E) Pathway enrichment analysis confirmed that interferon alpha and beta (type I) response was among the top downregulated pathways in the regressors.
Figure 3. Characterization of Cluster 1 reveals a subpopulation of monocytes primed for extravasation. A) GSEA showed a suppression of gene sets ‘regulation of endothelial cell proliferation’ ad ‘vasculature development’ in the regressors. Similarly, gene sets ‘regulation of type I interferon production’ and ‘type I interferon production’ were also suppressed in the regressors. B) The gene module score for the vasculature development gene set was calculated using the aggregated expression of the genes in this gene set. The gene module score for ‘Vasculature Development’-associated genes and ‘Endothelial Cell Proliferation’-related genes D) were both found to be suppressed in the regressors compared to other patient groups. E) Pathway enrichment analysis confirmed that interferon alpha and beta (type I) response was among the top downregulated pathways in the regressors.
To cite this abstract in AMA style:
Hofman A, Bearzi P, Pachera E, Bruni C, Li L, Much L, Bürki K, Becker M, Hoffmann-Vold A, Distler O. Multi-Omic Profiling Reveals a Monocyte-Vascular Signature Associated with the Regression of Skin Fibrosis in SSc [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/multi-omic-profiling-reveals-a-monocyte-vascular-signature-associated-with-the-regression-of-skin-fibrosis-in-ssc/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/multi-omic-profiling-reveals-a-monocyte-vascular-signature-associated-with-the-regression-of-skin-fibrosis-in-ssc/
