Session Information
Date: Monday, November 18, 2024
Title: Systemic Sclerosis & Related Disorders – Basic Science Poster II
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: The poor prognosis of patients with systemic sclerosis (SSc) underscores the need to prevent disease onset and irreversible organ damage. This study aims to investigate the molecular mechanisms underlying the very early stages of SSc before clinically evident fibrotic damage.
Methods: We recruited patients with very early SSc (veSSc, n=6), early established SSc (eSSc, n=7), and healthy controls (HC, n=6). veSSc was defined as previously described1, whereas eSSc patients met the 2013 ACR/EULAR criteria with a disease duration < 5 years. HCs were matched for age, sex, and ethnicity. Libraries of site-matched skin biopsies were prepared using the Chromium Next GEM single cell 3’ reagent kit v3.1 and sequenced. Raw reads were mapped to the human genome using Cellranger 7.1.0, and data analysis was performed using Seurat (5.1.0). Harmony was applied for integration. Speckle was used for cell type compositional analysis. Differential gene expression analysis (DGEA) was performed pairwise on both the pseudobulk (DESeq2) and single cell level (MAST). Over-representation analysis was conducted using the GO database in the clusterProfiler package.
- Muraru et al. (2024). “Arthritis in patients with very early systemic sclerosis: a comprehensive clinical and prognostic analysis.” Rheumatology.
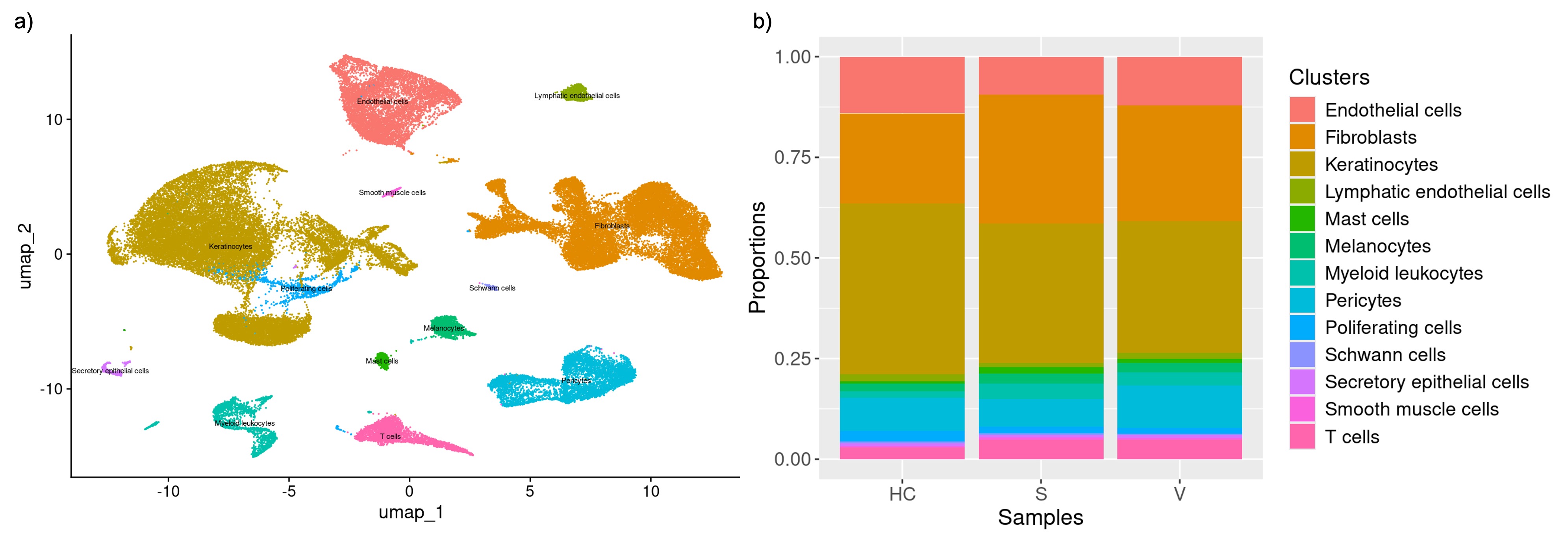
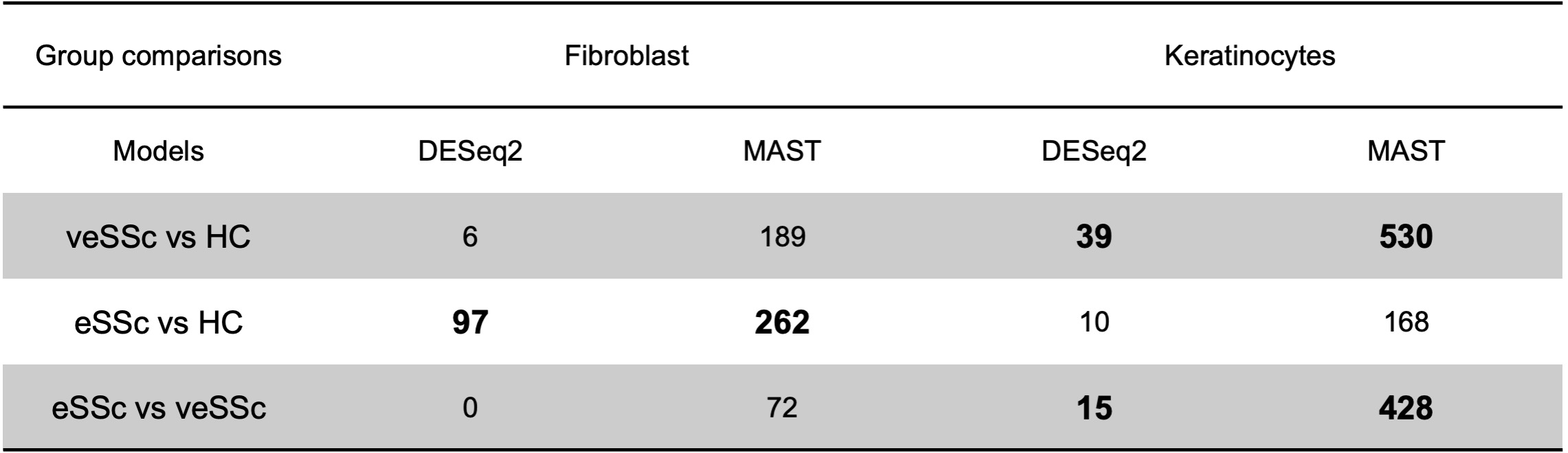
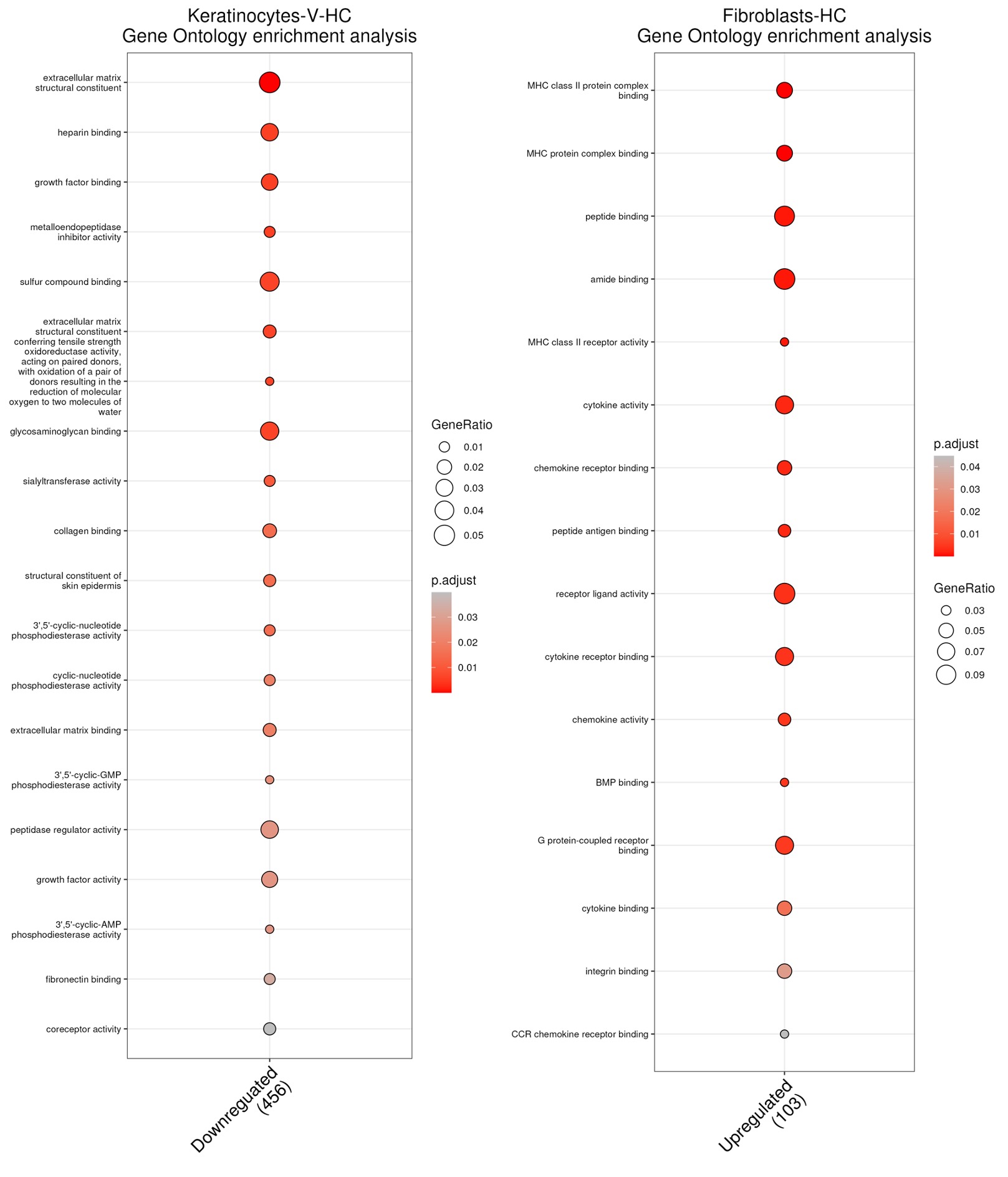
Results: Following quality control, we annotated a total of 63,844 cells into 13 cell types with keratinocytes (veSSc=7376, eSSc=8763, and HC=6853) and fibroblasts (veSSc=6460, eSSc=8073, and HC=3606) having the highest abundance (Fig.1a). Cell compositional analysis revealed that veSSc patients had already significantly different abundance compared to HC, and similar to eSSc (Fig.1b). In veSSc, we observed increased fibroblast (proportional ratio 1.28) and decreased keratinocyte abundance (proportional ratio 0.75). Pairwise DGEA for each cell type between all patient groups showed that keratinocytes had the highest number of differentially expressed genes (DEGs) in veSSc compared to HC with 39 significant DEGs deregulated in the DESeq2 model (|log2FC| >0.5, adjusted p-value < 0.05) and 530 in the MAST model (|log2FC| >1, adjusted p-value < 0.05) (Tab.1), while fibroblasts had fewer DEGs (6 in DESeq2 and 189 in MAST). Conversely, in eSSc vs. HC, fibroblasts displayed the most differentially expressed genes (97 in DESeq2 and 262 in MAST). Over-representation analysis of DEGs in veSSc keratinocytes (COL4A2, COL14A1, and COL5A2) indicated downregulation in extracellular matrix remodeling-related pathways (including “extracellular matrix structural constituent” and “collagen binding”). DEGs in veSSc fibroblasts (CD74, HLA-DRA, HLA-DPA1, and HLA-DRB1) showed upregulation in pro-inflammatory-related pathways (including “MHC class II protein complex binding” and “MHC class II receptor activity”) (Fig.2).
Conclusion: This is the first study providing a comprehensive molecular landscape of very early SSc. Keratinocytes display a prominent molecular gene dysregulation, while a proinflammatory signature is seen in fibroblasts. These observations have important implications for the very early pathogenesis of SSc.
To cite this abstract in AMA style:
Li L, Pachera E, Dobrota R, Bürki K, Mihai C, Elhai M, Much L, Hofman A, Bearzi P, Wagner S, Lötscher S, Hoffmann-Vold A, Distler O. Single-cell RNA Sequencing Profiling of Very Early-Stage Systemic Sclerosis Skin Reveals a Fibroblast Pro-inflammatory Gene Signature and Keratinocyte Dysregulation [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/single-cell-rna-sequencing-profiling-of-very-early-stage-systemic-sclerosis-skin-reveals-a-fibroblast-pro-inflammatory-gene-signature-and-keratinocyte-dysregulation/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-rna-sequencing-profiling-of-very-early-stage-systemic-sclerosis-skin-reveals-a-fibroblast-pro-inflammatory-gene-signature-and-keratinocyte-dysregulation/