Session Information
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Identification of disease and therapeutic biomarkers remains a barrier to the early diagnosis of and initiation of effective therapy for juvenile idiopathic arthritis (JIA). In this study, plasma metabolomic profiling was performed to identify disease-related metabolic biomarkers associated with JIA.
Methods: Plasma samples from treatment-naïve JIA patients and non-JIA reference patients were submitted to Metabolon, Inc. (Morrisville, North Carolina, USA) for global metabolomic profiling. The study was completed in a discovery and a validation cohort. The validation cohort consisted of 60 treatment-naïve JIA patients and 60 non-JIA reference patients that included patients undergoing endoscopy with either functional abdominal pain or Crohn’s disease. The independent validation cohort consisted of 52 treatment-naïve JIA patients and 38 non-JIA reference patients that were otherwise healthy. The datasets were analyzed individually and as a merged dataset. Univariate analysis was computed using MetaboAnalyst 5.0 and significant metabolites were identified based on a false-discovery rate adjusted p-value (q-value) of 0.05. Enrichment analysis based on the intersection of significant metabolites from the two cohorts was conducted by Chemical Similarity Enrichment Analysis (ChemRich) and a metabolic network map was created using MetaMapp and visualized using Cytoscape 3.9.1. Receiver operating characteristic (ROC) curve analysis was performed in MetaboAnalyst 5.0 to identify the top discriminating metabolite biomarkers based on the resulting area under the ROC curve (AUC).
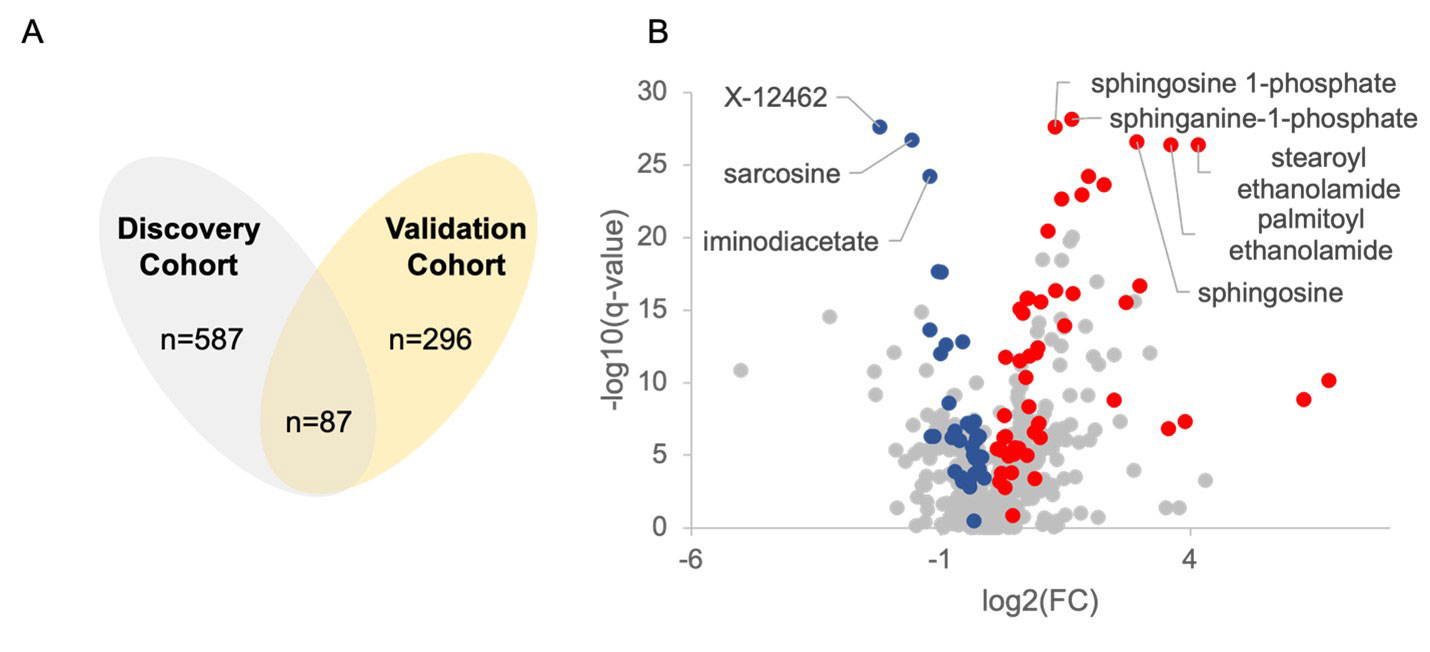
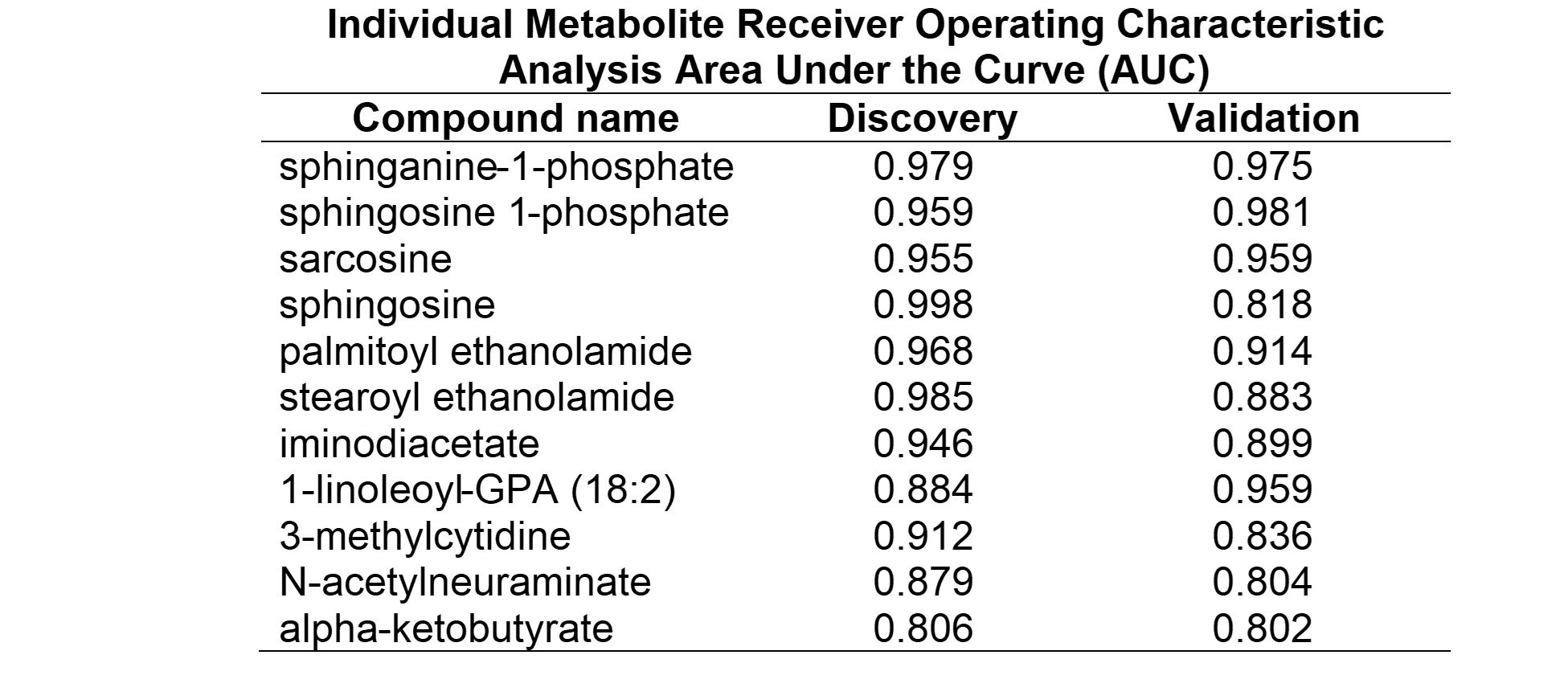
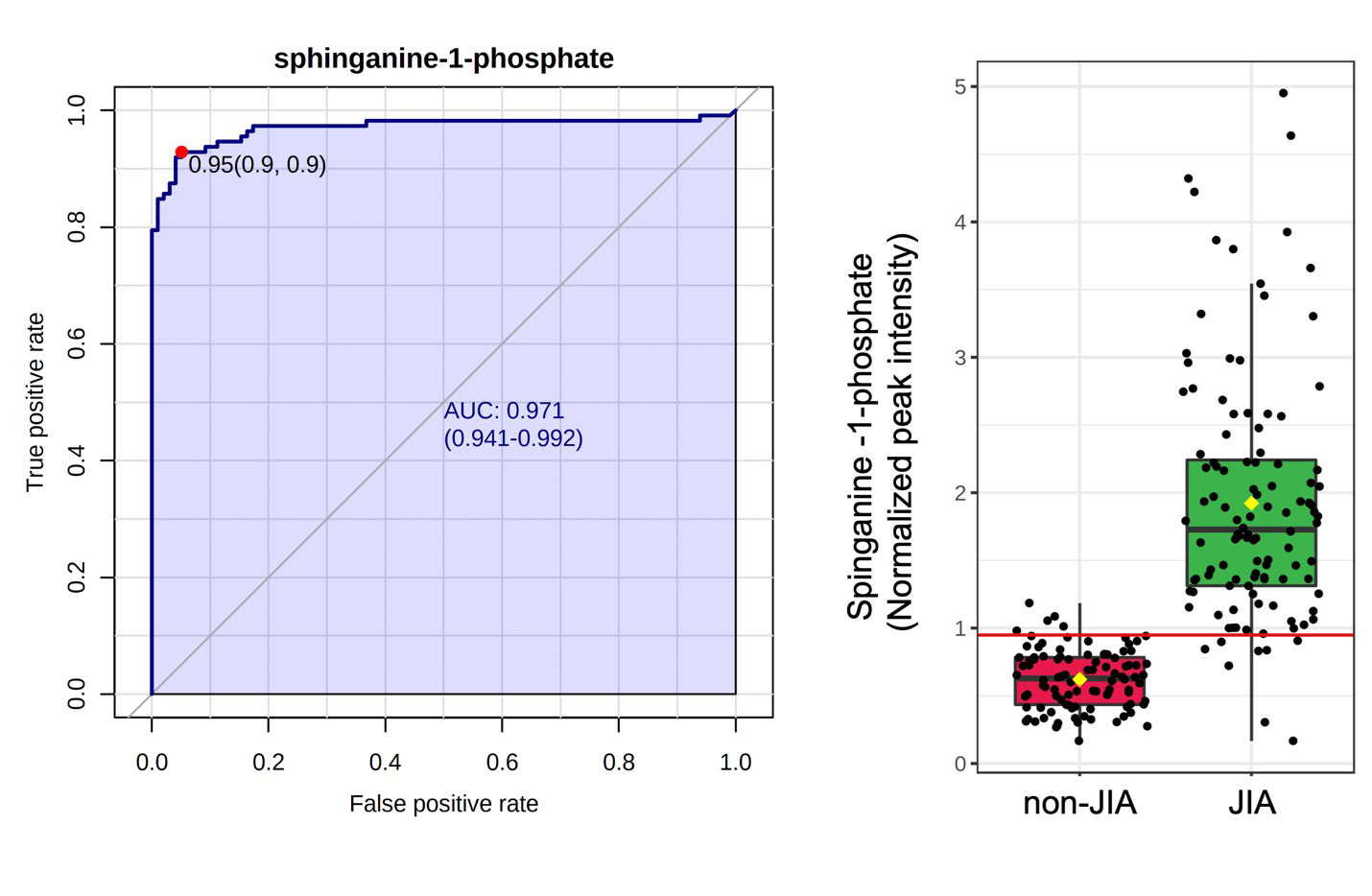
Results: A total of 869 metabolites were measured, comprised of 714 known and 155 unknown compounds. In the discovery cohort 587 metabolites were found to be significantly altered in JIA patients compared to the reference population (q< 0.05). In the validation cohort 296 metabolites were found to be significantly altered, with 87 overlapping metabolites demonstrating the same directional change in both cohorts (Figure 1A). JIA was associated with a notable increase in plasma levels of sphingosine metabolites and fatty acid ethanolamides, and decreased plasma levels of sarcosine, iminodiacetate, and the unknown metabolite X-12462 (Figure 1B). Chemical enrichment analysis identified cycloparaffins in the form of naproxen and its metabolites, unsaturated lysophospholipids, saturated phosphatidylcholines, sphingomyelins, ethanolamines and saturated ceramides as top discriminating biochemical clusters. ROC curve analysis identified 11 metabolites that would be classified as highly discriminatory based on an AUC > 0.80 (Table 1). The top discriminating metabolite identified was sphinganine-1-phosphate (AUC=0.97) (Figure 2).
Conclusion: Using an independent discovery and validation cohort approach, this study demonstrates metabolic differences in JIA compared to controls, and highlights the potential utility of plasma metabolomic profiling to identify disease biomarkers in JIA to guide accurate diagnosis and effective treatment. These findings suggest altered sphingosine metabolism in JIA resulting in accumulation of sphingosine and its immunomodulatory metabolites.
To cite this abstract in AMA style:
Kumar A, Tartarian J, Shakhnovich V, Langefeld C, Lovell D, Thompson S, Becker M, Funk R. Identification of Plasma Metabolomic Biomarkers of Juvenile Idiopathic Arthritis [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/identification-of-plasma-metabolomic-biomarkers-of-juvenile-idiopathic-arthritis/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-plasma-metabolomic-biomarkers-of-juvenile-idiopathic-arthritis/