Session Information
Date: Sunday, November 8, 2020
Session Type: Plenary Session
Session Time: 11:30AM-1:00PM
Background/Purpose: Checkpoint inhibitors (CI) used to treat cancer frequently trigger immune-related adverse events, including inflammatory arthritis. CI-related arthritis (CIrA) occurs in ~5% of treated patients, with clinical manifestations resembling rheumatoid arthritis (RA) and spondyloarthropathies (SpA). However, the cellular and molecular features of CIrA remain unknown, calling for a deeper understanding of its pathogenesis.
Methods: We conducted detailed immunophenotyping of synovial fluid (SF) mononuclear cells to compare CIrA, seropositive RA, and SpA using mass cytometry (CyTOF) (CIrA n=10, anti-PD-1-treated; 5 mono/oligoarthritis, 5 polyarthritis; RA n=11; SpA n=9). Significantly altered populations (p< 0.05) were identified using FlowSOM and validated by flow cytometry in additional SF and blood samples (CIrA n=15; RA n=15). SF CD8 T cell subpopulations were sorted for RNA-seq and intracellular staining. Transcriptomic features were recognized by identifying differentially expressed genes and by Gene Set Enrichment Analysis (GSEA) (q< 0.25). RA SF cells were cultured with cytokines (IFNα 1kU/mL or IFNγ 50ng/mL) and anti-CD3/CD28 for 72 hours and analyzed by flow cytometry.
Results: FlowSOM analysis of SF CyTOF revealed a CD38hiCD127– PD-1+ CD8 cell population uniquely expanded in CIrA (~25% of CD8 in CIrA, a 3.4-fold increase over RA/SpA), which was confirmed in additional SF samples by flow cytometry. In contrast, T cells with the converse CD38–CD127+ phenotype were reduced in CIrA compared to RA/SpA. CD38hiCD127– CD8 cells were also expanded in blood of CIrA patients (5% of CD8 in CIrA, a 2.8-fold increase over RA) (Fig. 1).
RNA-seq analysis of the expanded CD38hiCD127– subset revealed significantly increased expression of inflammatory and cytotoxic molecules including granzyme A/B/H/K, perforin and IFNγ, with expression levels of these genes comparable to those in sorted KLRG1+ cytotoxic T cells. Flow cytometry of ex vivo stimulated T cells confirmed a higher frequency of granzyme B+ and perforin+ cells in the CD38hiCD127- population (Fig. 2). GSEA revealed an increased proliferation signature in CD38hiCD127– cells compared to CD38–CD127+ cells, with upregulation of genes involved in DNA replication, mitosis and cell division (q< 0.1, FC >1.5 over CD38–CD127+ cells). This feature was not shared by PD1– cells or KLRG1+ cells (Fig 3). CyTOF confirmed >40% of CD38hi CD127– PD-1+ CD8 T cells expressed Ki67. GSEA of SF T cells also revealed a marked enrichment of IFN-inducible genes in CIrA compared to SpA/RA. Treatment of RA SF T cells with IFNα, but not IFNγ, induced CD8 T cells to acquire a CD38hiCD127– phenotype resembling that seen in CIrA (Fig. 3).
Conclusion: CyTOF analysis of SF mononuclear cells revealed a highly expanded PD-1+CD38hiCD127– CD8 T cell population in CIrA that is not shared by RA or SpA. This T cell subset shows cytotoxic and proliferative features at both the gene and protein expression level. CD38hiCD127– CD8 T cells in CIrA samples showed an IFN signature, and treatment of T cells with IFNα induced this phenotype in vitro. This work reveals a unique T cell phenotype in CIrA and suggests type I IFN may be a pathologic driver in this condition.
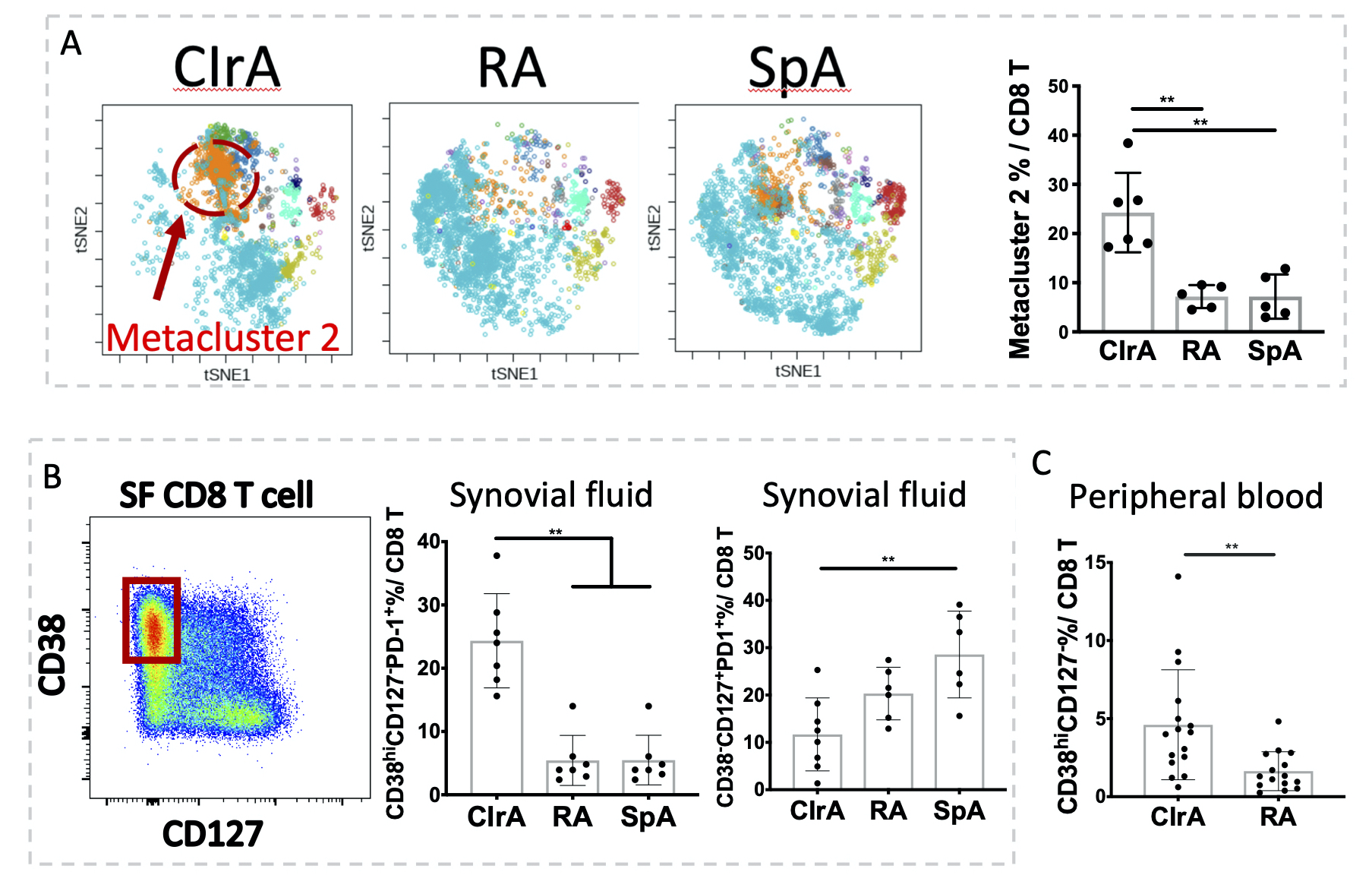 Figure 1. FlowSOM analysis of CD8 T cells revealed the expansion of a PD-1+CD38hiCD127- population in synovial fluid and peripheral blood of checkpoint inhibitor-related arthritis (CIrA). A) FlowSOM analysis of CyTOF CD8 T cells highlighted an expanded metacluster 2 in CIrA SF (Kruskal-Wallis test, CIrA n=6, RA/SpA n=5). B) Biaxial gating of flow cytometry data confirmed expanded CD38hiCD127-PD1+ cells and reduced CD38-CD127+ CD8 T cells in CIrA SF (Kruskal-Wallis test, n=7). C) CD38hiCD127- CD8 cells are expanded in blood of CIrA patients (t-test, n=15). Mean±SD shown, **p < 0.01.
Figure 1. FlowSOM analysis of CD8 T cells revealed the expansion of a PD-1+CD38hiCD127- population in synovial fluid and peripheral blood of checkpoint inhibitor-related arthritis (CIrA). A) FlowSOM analysis of CyTOF CD8 T cells highlighted an expanded metacluster 2 in CIrA SF (Kruskal-Wallis test, CIrA n=6, RA/SpA n=5). B) Biaxial gating of flow cytometry data confirmed expanded CD38hiCD127-PD1+ cells and reduced CD38-CD127+ CD8 T cells in CIrA SF (Kruskal-Wallis test, n=7). C) CD38hiCD127- CD8 cells are expanded in blood of CIrA patients (t-test, n=15). Mean±SD shown, **p < 0.01.
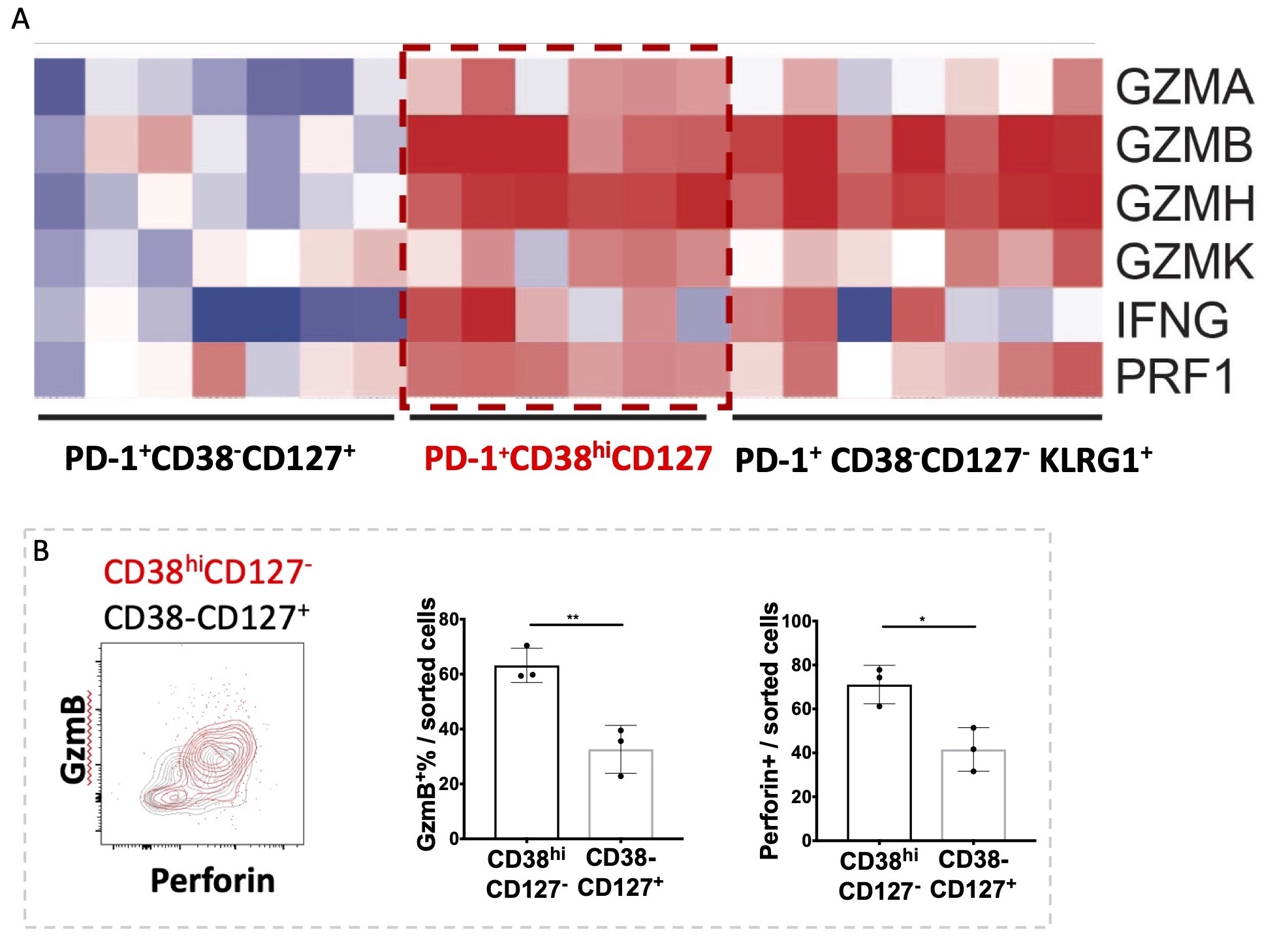 Figure 2. RNA-seq analysis and flow cytometry staining of CD38hiCD127- CD8 T cells in CIrA SF demonstrated cytotoxic and inflammatory features. A) Heatmap of cytotoxic molecules enriched in CD38hiCD127- cells. B) Enrichment of intracellular granzyme B and perforin in sorted CD38hiCD127- CD8 T cells. Mean±SD shown, *p < 0.05, **p < 0.01.
Figure 2. RNA-seq analysis and flow cytometry staining of CD38hiCD127- CD8 T cells in CIrA SF demonstrated cytotoxic and inflammatory features. A) Heatmap of cytotoxic molecules enriched in CD38hiCD127- cells. B) Enrichment of intracellular granzyme B and perforin in sorted CD38hiCD127- CD8 T cells. Mean±SD shown, *p < 0.05, **p < 0.01.
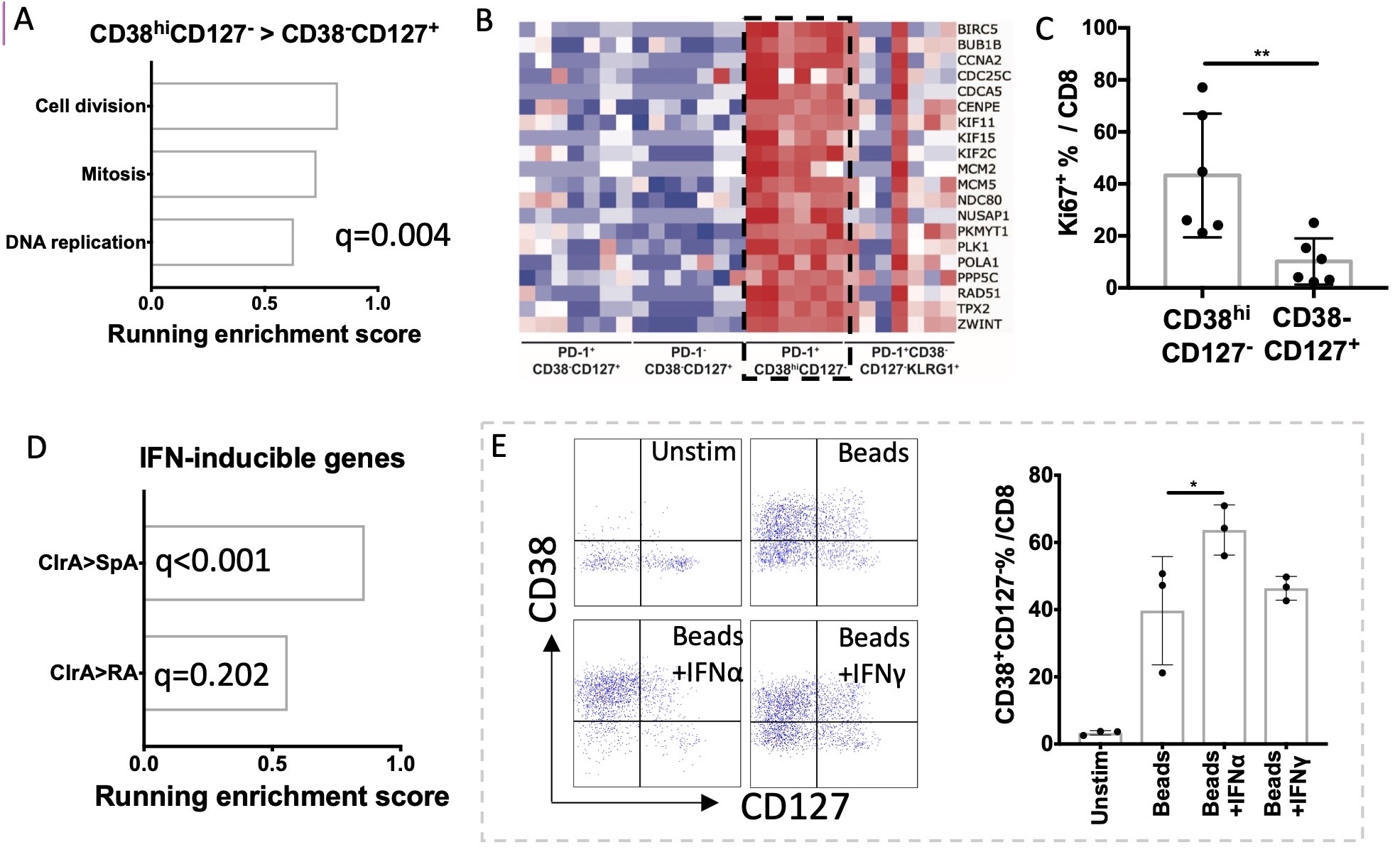 Figure 3. CD38hiCD127- CD8 T cells in CIrA SF showed a proliferation feature, and a phenotype induced by IFNα. A) GSEA identified enrichment of proliferation gene sets in CD38hiCD127- CD8 T cells; B) Heatmap of proliferation genes enriched in CD38hiCD127- CD8 T cells. C) CyTOF showed increased expression of Ki67 in CD38hiCD127- CD8 T cells. D) GSEA showed an IFN signature in CIrA cells. E) RA SF cells were cultured unstimulated or stimulated with anti-CD3/28 activation beads with or without IFNα or IFNγ as indicated for 72 hours. CD38hiCD127- phenotype resembling that seen in CIrA was induced by IFNα but not IFNγ. Mean±SD shown, *p < 0.05, **p < 0.01.
Figure 3. CD38hiCD127- CD8 T cells in CIrA SF showed a proliferation feature, and a phenotype induced by IFNα. A) GSEA identified enrichment of proliferation gene sets in CD38hiCD127- CD8 T cells; B) Heatmap of proliferation genes enriched in CD38hiCD127- CD8 T cells. C) CyTOF showed increased expression of Ki67 in CD38hiCD127- CD8 T cells. D) GSEA showed an IFN signature in CIrA cells. E) RA SF cells were cultured unstimulated or stimulated with anti-CD3/28 activation beads with or without IFNα or IFNγ as indicated for 72 hours. CD38hiCD127- phenotype resembling that seen in CIrA was induced by IFNα but not IFNγ. Mean±SD shown, *p < 0.05, **p < 0.01.
To cite this abstract in AMA style:
Wang R, Chan K, Cunningham-Bussel A, Vitone G, Tirpack A, Benson C, Keras G, Jonsson A, Brenner M, Donlin L, Bass A, Rao D. High-dimensional Analyses of Checkpoint-inhibitor Related Arthritis Synovial Fluid Cells Reveal a Unique, Proliferating CD38hi Cytotoxic CD8 T Cell Population Induced by Type I IFN [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/high-dimensional-analyses-of-checkpoint-inhibitor-related-arthritis-synovial-fluid-cells-reveal-a-unique-proliferating-cd38hi-cytotoxic-cd8-t-cell-population-induced-by-type-i-ifn/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/high-dimensional-analyses-of-checkpoint-inhibitor-related-arthritis-synovial-fluid-cells-reveal-a-unique-proliferating-cd38hi-cytotoxic-cd8-t-cell-population-induced-by-type-i-ifn/
