Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Cognitive impairment (CI) is a debilitating, untreatable problem for up to 80% of individuals with SLE and human and mouse data suggest multiple SLE-related pathogenic mechanisms. However, objective diagnosis of CI with attribution to SLE-mediated mechanisms is difficult, given the limitations of cognitive testing and confounding influences of medications and comorbid disease. We have previously identified hypermetabolic brain regions in SLE subjects that correlate with poor cognitive performance.1 We analyzed FDG PET scans from SLE subjects using deep learning (DL), a form of artificial intelligence, to identify a specific brain network (SLE-CogNet) associated with CI in SLE patients.
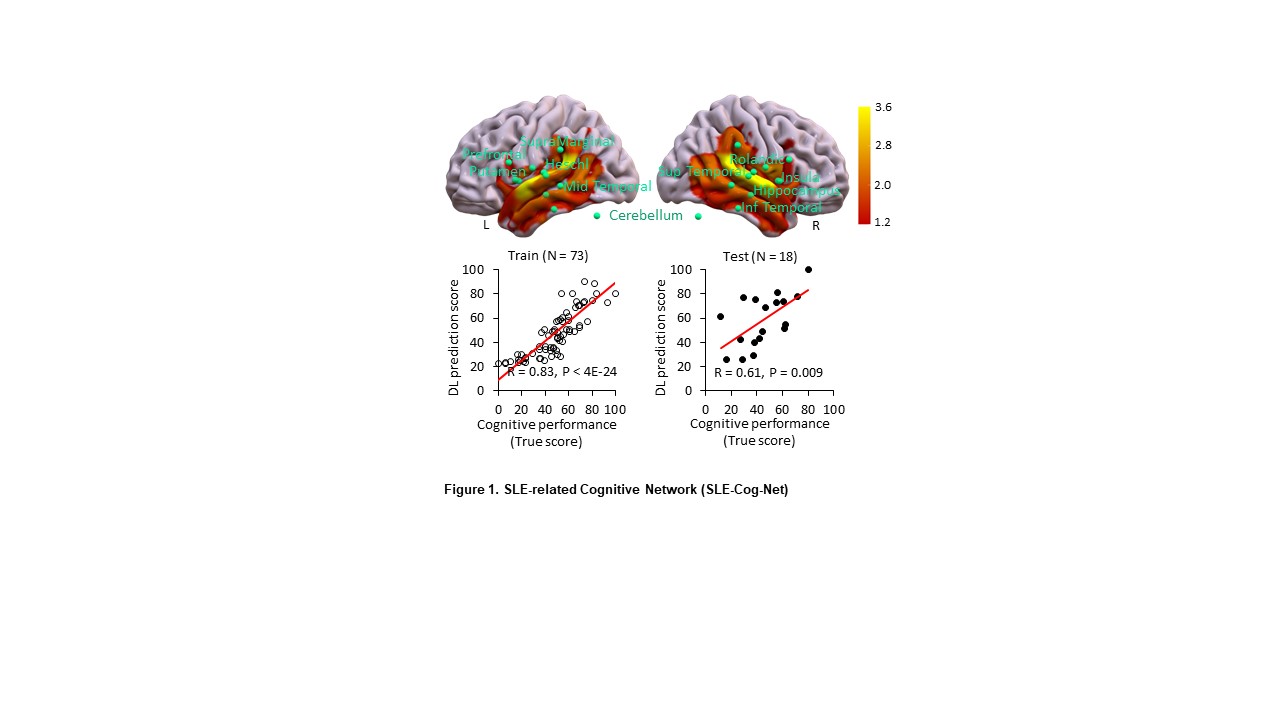
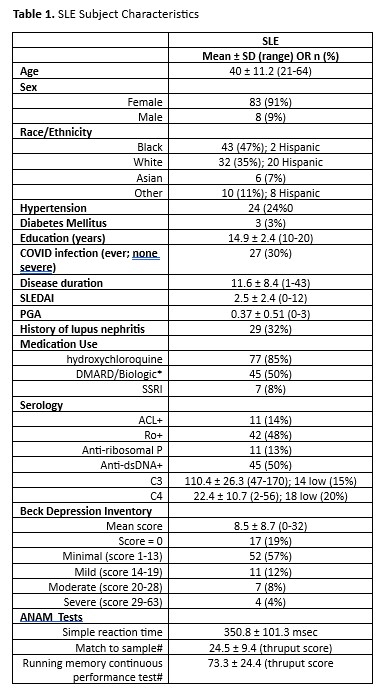
Methods: DL methods were applied to FDG PET data from 91 SLE subjects with concurrent Automated Neuropsychological Assessment Metrics (ANAM) test data. Subjects had stable disease activity, were on prednisone doses ≤ 10 mg, and had no history of CNS disease of any kind. We implemented a 3-dimensional (3D) convolutional neural network (CNN) based on a residual neural network algorithm (ResNet101).2 The cohort was divided randomly into training (n=73: 80%) and testing (n=18: 20%) sets. The 3D CNN was trained on individual FDG PET images and corresponding composite ANAM test scores to identify a significant SLE-CogNet that was validated in the respective testing set. Following training, we employed gradient-weighted class activation mapping (Grad-CAM)3 to generate an explainable map for each scan. DL expression scores for individual SLE subjects were computed across the entire network. For post-hoc analyses, a DL prediction score was determined for 95 regions of interest (ROI) from the automated anatomical atlas as the mean value for the relevant regions.
Results: The resulting SLE-CogNet is characterized by contributions from the hippocampus, prefrontal cortex, putamen, insula, temporal cortex, supramarginal gyrus, rolandic operculum, and cerebellum (Fig. 1). Composite ANAM scores correlated significantly with individual subject DL expression scores for SLE-CogNet in both the training (r=0.83, p< 0.001) and testing (r=0.61, p=0.009) sets. Compared to ANAM data from age and sex matched healthy individuals (n=72), SLE mean ANAM scores were significantly lower (p < 0.01). The R2 measure of the DL model was 69%. Post hoc analyses show significant correlations between C3, anti-dsDNA antibody titers and DL expression scores in multiple regions (p≤0.04).
Conclusion: DL analysis of FDG PET data identifies a candidate SLE-CogNet, composed mainly of hypermetabolic regions associated with CI in previous SLE studies.1 This network may provide an objective measure of CI in SLE that can be used to monitor patients and assess response to treatment.
1 M. Mackay et al., Metabolic and microstructural alterations in the SLE brain correlate with cognitive impairment. JCI Insight 4 (2019)
2 He K et al. Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition. (2015)
3 R. Selvaraju et al. Visual Explanations from Deep Networks via Gradient-Based Localization. Int J Comput Vis 128 (2016)
To cite this abstract in AMA style:
Nguyen N, Vo A, Tang C, Anderson E, Aranow C, Diamond B, Eidelberg D, Mackay M. Using FDG PET Brain Scans and Deep Learning Analyses to Identify a Specific Network Correlated with Cognitive Impairment in SLE [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/using-fdg-pet-brain-scans-and-deep-learning-analyses-to-identify-a-specific-network-correlated-with-cognitive-impairment-in-sle/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/using-fdg-pet-brain-scans-and-deep-learning-analyses-to-identify-a-specific-network-correlated-with-cognitive-impairment-in-sle/