Session Information
Session Type: Plenary Session
Session Time: 11:30AM-1:00PM
Background/Purpose: Treatment of lupus nephritis relies on renal histopathological features. However, renal biopsies do not capture patient-specific active biological pathways. Urine proteomic biomarkers could revolutionize the diagnosis and management of lupus nephritis by predicting active intrarenal biological pathways and can be noninvasively monitored over time.
Methods: One thousand proteins were quantified (RayBiotech) in a total of 112 longitudinal urine samples from 30 SLE patients with active lupus nephritis and 7 healthy controls (HC). The proteins and molecular pathways detected in the urine proteome at the time of biopsy were then analyzed with respect to lupus nephritis class, response to treatment after 1 year, histopathological features (activity and chronicity indeces), and trajectory over time (baseline and week 12, 26, and 52). The intrarenal expression of candidate biomarkers was evaluated using single cell transcriptomics of renal biopsies from patients with active lupus nephritis.
Results: There were 237 proteins (FDR < 10%) enriched in the urine of patients with lupus nephritis reflecting several molecular pathways involving chemotaxis, extracellular matrix remodeling, and activation of neutrophils and platelets. Hierarchical clustering using urine proteomics segregated SLE patients into 2 groups, with 80% of complete responders clustering together. This finding could not be similarly reproduced using standard features including baseline proteinuria, creatinine, histologic activity or chronicity scores, or class, indicating unique informative features of urine proteomics (Fig. 1). Patients with proliferative lupus nephritis (class III or IV) had stronger activation of chemotaxis pathways. IL-16 was the urinary protein most significantly increased in proliferative disease compared to membranous (FC 6, p=0.002) (Fig. 2A). Assessment of urine proteins that correlated with histologic activity kidney highlighted IL-16 as the single most strongly correlated protein with histologic activity (r=0.69, p=9.5∙10-5; Fig. 2B). IL-16 concentration was independent of the amount of proteinuria and progressively diminished over time in patients who were responding to immunosuppression (Fig. 2C). Single cell RNA sequencing revealed significant intrarenal expression of IL16 by all infiltrating immune cells and highlighted IL16 as the second most expressed cytokine in lupus nephritis kidneys out of a compendium of 236 cytokines (Fig. 3A-B).
Conclusion: Urine proteomics can noninvasively identify active and biologically relevant pathways in lupus nephritis. Integrated urine proteomics and renal single cell transcriptomics revealed that IL-16, a CD4 ligand with chemotactic and proinflammatory functions, was one of the most expressed cytokine in lupus nephritis. As a urine proteomic biomarker, IL-16 may predict renal histological activity and could be monitored over time to assess response to immunosuppression. Urinary IL-16 is independent of proteinuria thus potentially providing actionable clinical information that is not captured by currently used biomarkers. Further studies are ongoing to validate these findings.
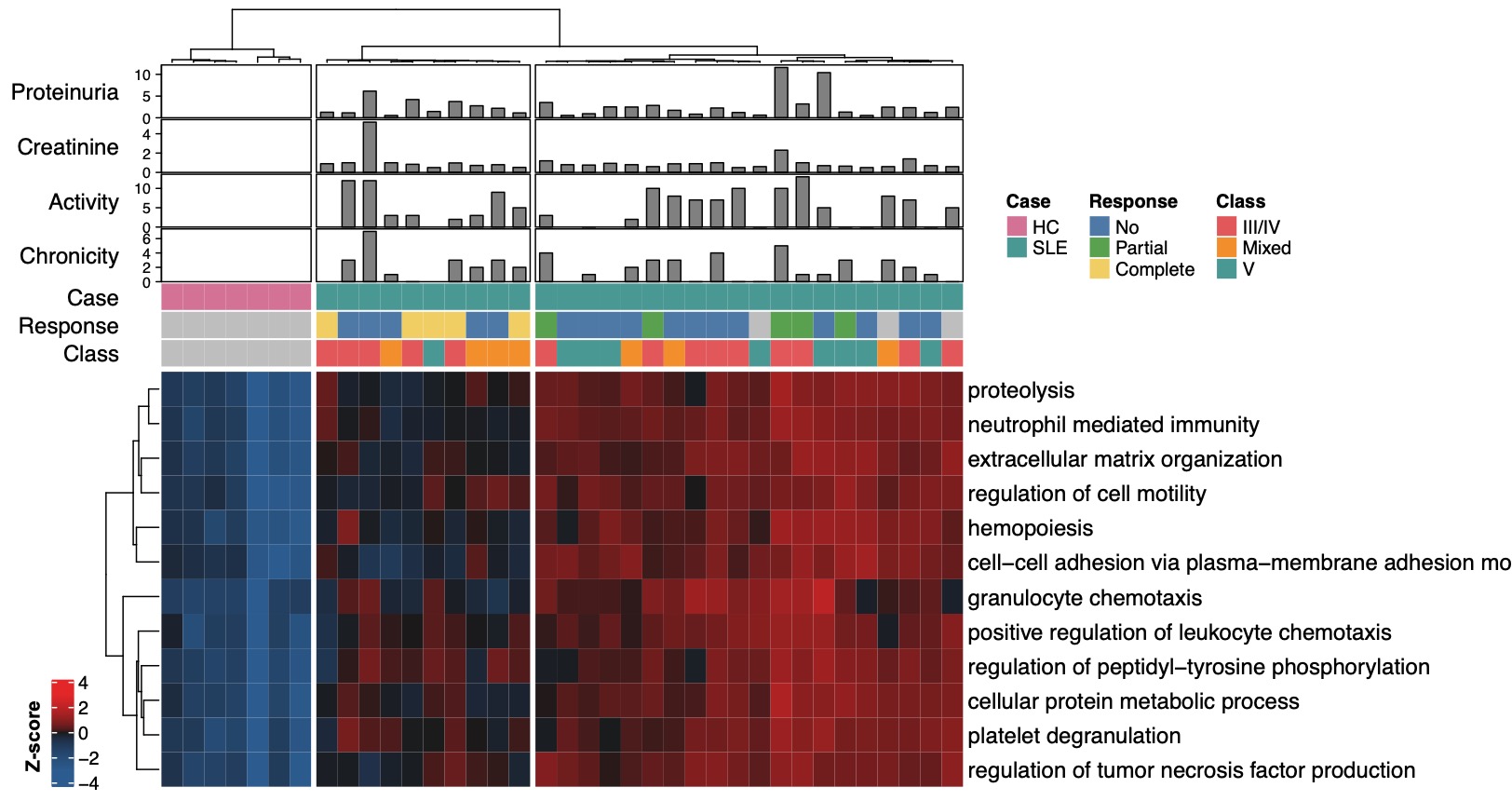 Figure 1. Concentration of enriched proteomic pathways in lupus neprhitis urine. The relative concentration of the 12 non overlapping pathways (GO Biological Process) enriched in the urine of lupus nephritis (LN) patients is reported in this heatmap. Colors represent row-normalized Z-scores. Unsupervised hierarchical clustering identified the healthy controls and two clusters of LN patients. The LN cluster on the left showed overall lower concentration of proteins in the 12 pathways and included 4/5 of complete responders.
Figure 1. Concentration of enriched proteomic pathways in lupus neprhitis urine. The relative concentration of the 12 non overlapping pathways (GO Biological Process) enriched in the urine of lupus nephritis (LN) patients is reported in this heatmap. Colors represent row-normalized Z-scores. Unsupervised hierarchical clustering identified the healthy controls and two clusters of LN patients. The LN cluster on the left showed overall lower concentration of proteins in the 12 pathways and included 4/5 of complete responders.
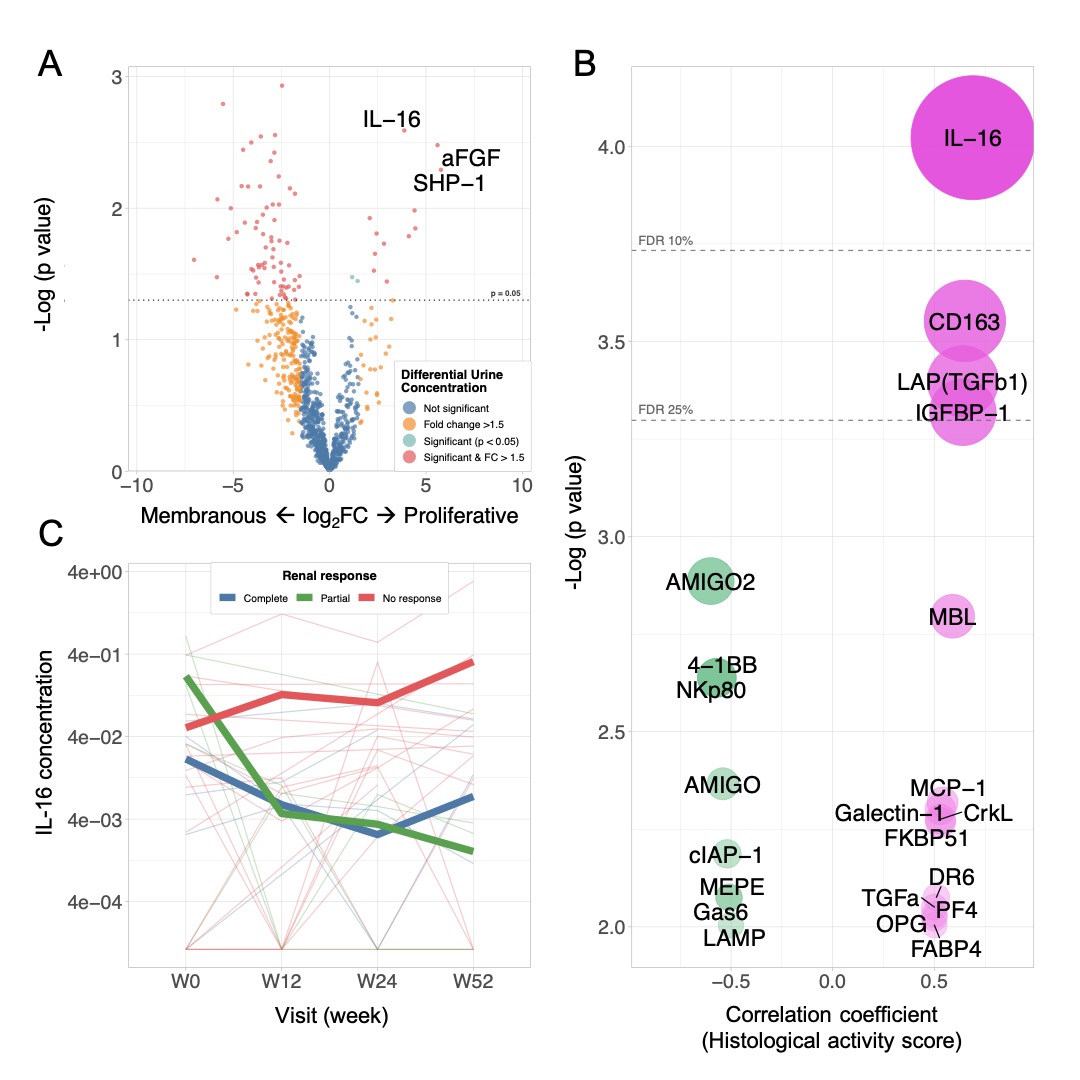 Figure 2. Urinary IL-16 is correlated with renal histological activity and response to treatment. (A) Volcano plot comparing the concentration of 1000 urinary proteins in proliferative vs pure membranous lupus nephritis. (B) Top 20 Pearson’s correlations of urinary proteins with renal histological NIH activity index. IL-16 was the only protein with a false discovery rate (FDR) < 10%. Circle areas represent a combination of the absolute correlation coefficient and p value. (C) Longitudinal trends of urinary IL-16 at baseline, week 12, 24 and 52. Thick lines represent the mean concentration according to the response status at week 52.
Figure 2. Urinary IL-16 is correlated with renal histological activity and response to treatment. (A) Volcano plot comparing the concentration of 1000 urinary proteins in proliferative vs pure membranous lupus nephritis. (B) Top 20 Pearson’s correlations of urinary proteins with renal histological NIH activity index. IL-16 was the only protein with a false discovery rate (FDR) < 10%. Circle areas represent a combination of the absolute correlation coefficient and p value. (C) Longitudinal trends of urinary IL-16 at baseline, week 12, 24 and 52. Thick lines represent the mean concentration according to the response status at week 52.
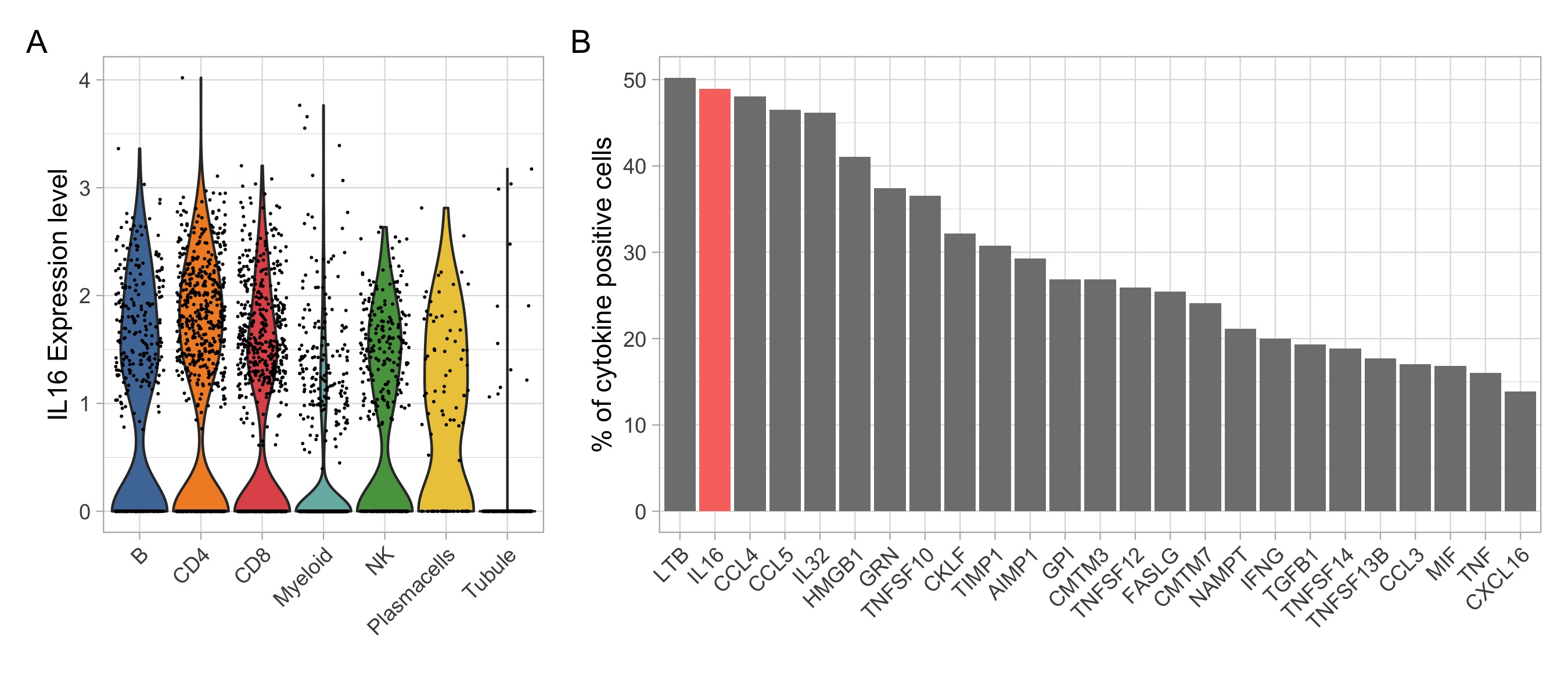 Figure 3. Intrarenal expression of IL16 based on single cell RNA sequencing of lupus nephritis kidney biopsies. (A) Violin plots showing IL16 expression by all kidney infiltrating immune cells. (B) Percentage of cytokine positive cells across all kidney infiltrating immune cells. The bar plot shows the top 25 cytokines out of a comprehensive list of 236 obtained from the “Cytokine Registry” (Immport) and the Gene Ontology database.
Figure 3. Intrarenal expression of IL16 based on single cell RNA sequencing of lupus nephritis kidney biopsies. (A) Violin plots showing IL16 expression by all kidney infiltrating immune cells. (B) Percentage of cytokine positive cells across all kidney infiltrating immune cells. The bar plot shows the top 25 cytokines out of a comprehensive list of 236 obtained from the “Cytokine Registry” (Immport) and the Gene Ontology database.
To cite this abstract in AMA style:
Fava A, Buyon J, Mohan C, Zhang T, Belmont H, Izmirly P, Clancy R, Monroy-Trujillo J, Berthier C, Davidson A, Hacohen N, Wofsy D, Rao D, Raychaudhuri S, Accelerating Medicines Partnership in SLE Network T, Apruzzese W, Petri M. Urine Proteomics and Single Cell Transcriptomics Identify IL-16 as a Biomarker for Lupus Nephritis [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/urine-proteomics-and-single-cell-transcriptomics-identify-il-16-as-a-biomarker-for-lupus-nephritis/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/urine-proteomics-and-single-cell-transcriptomics-identify-il-16-as-a-biomarker-for-lupus-nephritis/
