Session Information
Session Type: Poster Session A
Session Time: 8:30AM-10:30AM
Background/Purpose: Osteoarthritis (OA) is a chronic disabling disease, for which there are only limited treatment options. One major challenge in the development of effective treatment strategies for knee OA is the complexity of the underlying pathogenesis. Segmenting patient populations using readily available clinical, radiographic and biological features to define different phenotypes is a prerequisite for personalized patient management. The present study aimed to identify clinical OA phenotypes and their progression based on readily available characteristics using two fundamentally different machine learning algorithms.
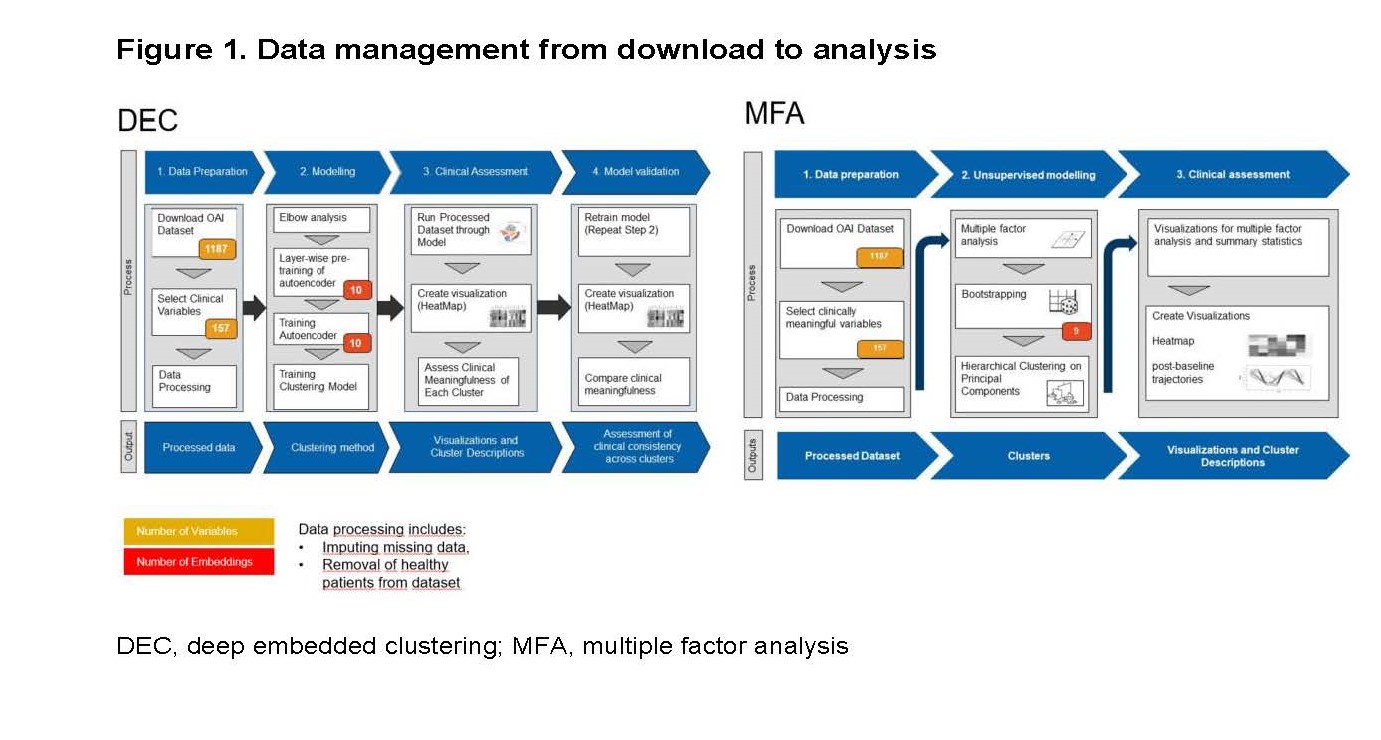
Methods: Clusters were based on 157 easily accessible clinical variables collected from the Osteoarthritis Initiative (OAI) baseline dataset, which included 4,674 participants covering the entire spectrum of knee OA severity. After reduction of data-dimensionality (i.e., reduction of the complexity without loss of information), the analysis relied on deep embedded clustering (DEC) and multiple factor analysis (MFA), respectively, for the identification of homogenous groups (Figure 1). In addition, radiographic progression of OA was analyzed in the different clusters both clinically (using the Knee Injury and Osteoarthritis Outcome Score, KOOS) over 9 years and structurally (via minimal medial joint space width) over 4 years using a linear mixed effects model.
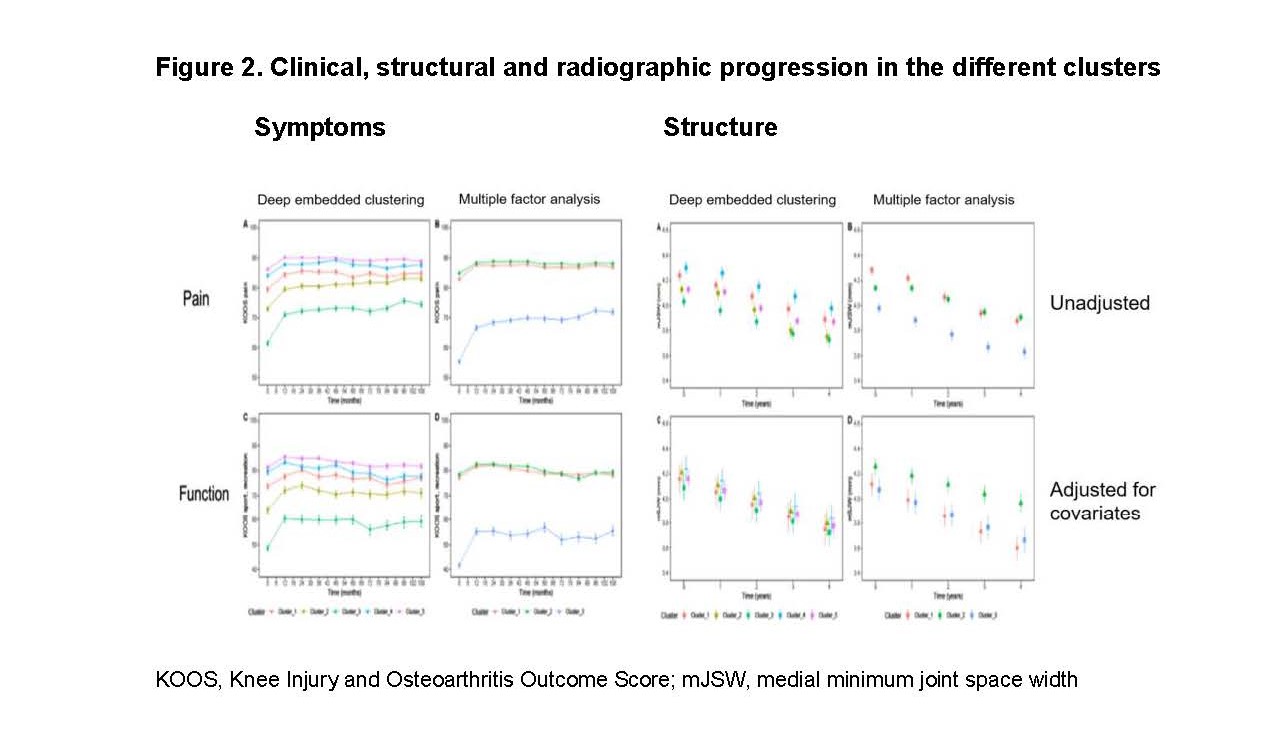
Results: The analysis yielded 5 clusters in DEC and 3 clusters in MFA with overlap between the two techniques: Both the approaches identified a comorbid cluster which comprised predominantly females with high body mass index (BMI), peri-articular pain, a high burden of comorbidities and depression, low activity levels, pain (KOOS) and functional limitations (cluster 3 in both analyses, Figure 2). Moreover, both approaches identified a cluster of more active participants with low levels of pain measured with the Physical Activity Scale for the Elderly (PASE) and KOOS (cluster 1, Figure 2). This cluster was characterized by a dominance of male patients in MFA analysis. The 3rd MFA cluster was slightly older, relatively lean, with low pain but functional limitations (cluster 2). The three remaining clusters after DEC differed in their levels of pain, waist circumference, activity and/or age. After an initial improvement in all clusters, the intensity of symptoms was stable on cluster-specific level (Figure 2).
Radiographic progression profiles differed only for MFA clusters (Figure 2): the more active (cluster 1), as well as the comorbid cluster (cluster 3) showed faster progression than the older, less active patients (cluster 2). Age, gender, BMI, abdominal obesity, presence of knee pain and malalignment were independent risk factors for structural progression.
Conclusion: We identified differentiated, clinically relevant subpopulations within the OAI data set by using two different machine learning approaches. The cluster-specific baseline characteristics will allow for refinement of inclusion criteria for future interventional clinical trials in specific knee OA phenotypes.
To cite this abstract in AMA style:
Demanse D, Tankó L, Lustenberger P, Nikolaus P, Rasin I, Brennan D, Saxer F, Roubenoff R, Premji S, Conaghan P, Schieker M. Unsupervised Machine-learning Algorithms for the Identification of Clinical Phenotypes in the Osteoarthritis Initiative Database [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/unsupervised-machine-learning-algorithms-for-the-identification-of-clinical-phenotypes-in-the-osteoarthritis-initiative-database/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/unsupervised-machine-learning-algorithms-for-the-identification-of-clinical-phenotypes-in-the-osteoarthritis-initiative-database/