Session Information
Date: Monday, November 8, 2021
Session Type: Poster Session C
Session Time: 8:30AM-10:30AM
Background/Purpose: Single biomarkers have limited utility to date in guiding clinical care in RA. There is growing interest in applying machine learning algorithms to combine demographic, clinical, and biomarker data to better identify and stratify RA patient outcomes. We aimed to determine if unsupervised machine learning methods can be employed in a racially and ethnically diverse RA cohort to identify clusters of patients with different disease activity trajectories, as measured by DAS28ESR.
Methods: Data are derived from the longitudinal, observational University of California, San Francisco RA Cohort from years 2011-2018. Along with routine labs, medication use and disease activity assessments, a multiple biomarker of disease activity (MBDA) panel was obtained at each clinical encounter. The MBDA measures 12 unique serum biomarkers. All observations were collapsed to create a cross-sectional dataset before clustering. Missing data were imputed using multiple imputation with chained equations. Data were standardized in preparation for clustering. Patient clusters were identified by unsupervised K-prototype clustering. Longitudinal disease activity (DAS28ESR) trajectories and 95%CIs were plotted for each cluster. Lasso regression was used to identify biomarkers independently associated with DAS28ESR within the whole cohort and by cluster.
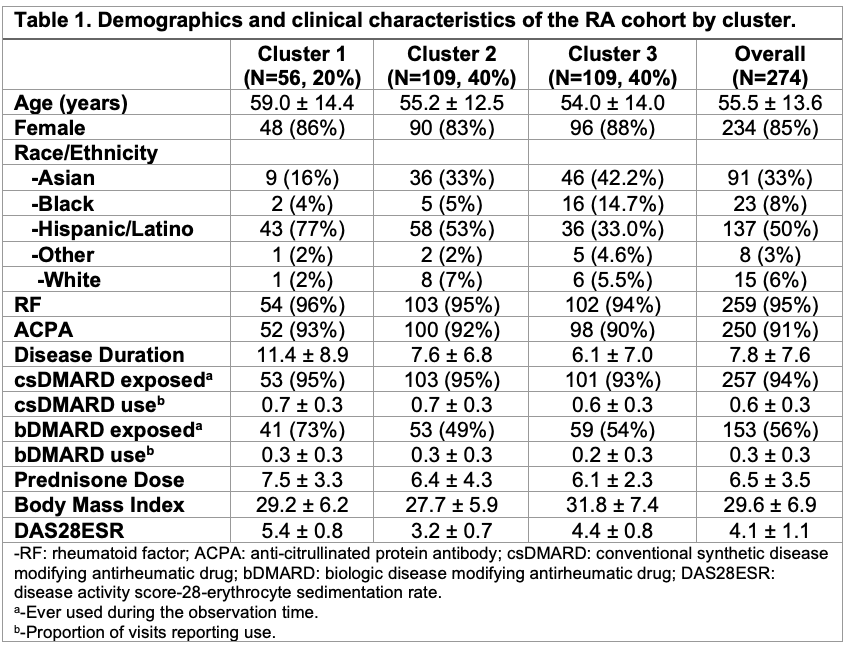
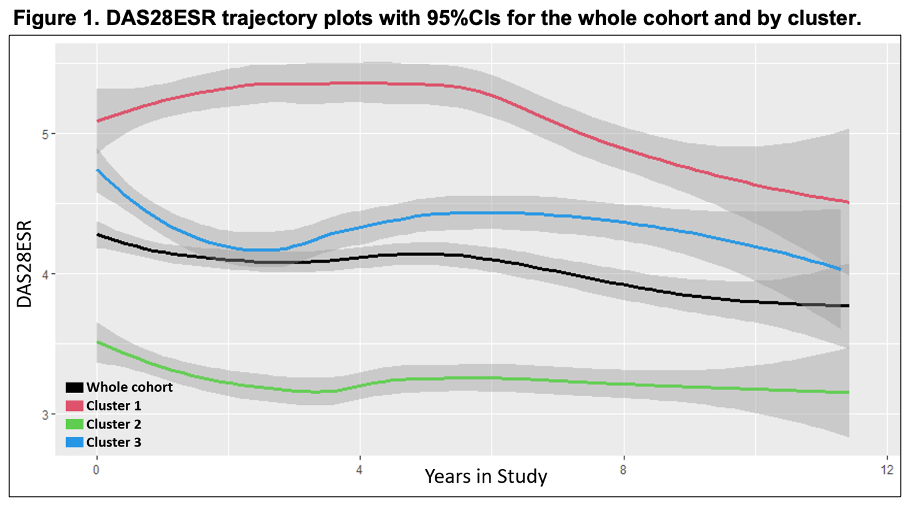
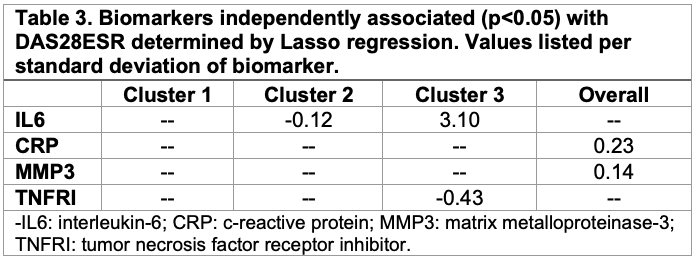
Results: Three distinct clusters were identified in our dataset. Cluster 1 (C1) was our smallest cluster (N=56, 20%), with the oldest age (59±14.4), highest proportion of Hispanic/Latino participants (n=43, 77%), the longest disease duration (11.4±8.9) and highest proportion of biological DMARD exposure (N=41, 73%) (Table 1). C1 also had the highest disease activity measured by DAS28ESR of 5.4±0.8. C2 and C3 both had 109 (40%) participants with similar ages 55.2±12.5 and 54.0±14.0, respectively. C3 had the highest proportion of Asian participants of the clusters (N=91, 33%) and the highest BMI of the cohort at 31.8±7.4. Notably, C2 had the lowest DAS28ESR of 3.2±0.7 (Figure 1). C1 had the highest mean DAS28ESR trajectory over time, whereas C3 had high disease activity that decreased over time. C2 had the lowest disease activity throughout the observation period. CRP and matrix metalproteinase-3 (MMP3) both had significant positive associations with DAS28ESR in our lasso regression model of the whole cohort (Table 2). No significant biomarker associations with DAS28ESR were found in C1. IL-6 had a negative association with DAS28ESR in C2 whereas IL-6 had a positive association and TNF-receptor inhibitor had a negative association with DAS28ESR in C3.
Conclusion: Using machine learning methods, we identified 3 clusters of patients in a racially and ethnically diverse longitudinal RA cohort. Each cluster had distinct disease activity trajectories and biomarker associations. This project demonstrated that machine learning methods can be applied to a moderate size RA cohort. Future work will be focused on evaluating baseline data to predict disease activity overtime and validating our findings in an external cohort.
To cite this abstract in AMA style:
Lui G, Singh N, Andrews J, Graf J, Wysham K. Unsupervised Clustering Identifies Unique Subsets of Patients in a Racially and Ethnically Diverse Rheumatoid Arthritis Cohort [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/unsupervised-clustering-identifies-unique-subsets-of-patients-in-a-racially-and-ethnically-diverse-rheumatoid-arthritis-cohort/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/unsupervised-clustering-identifies-unique-subsets-of-patients-in-a-racially-and-ethnically-diverse-rheumatoid-arthritis-cohort/