Session Information
Date: Sunday, November 12, 2023
Title: Abstracts: Systemic Sclerosis & Related Disorders I: Translational
Session Type: Abstract Session
Session Time: 2:00PM-3:30PM
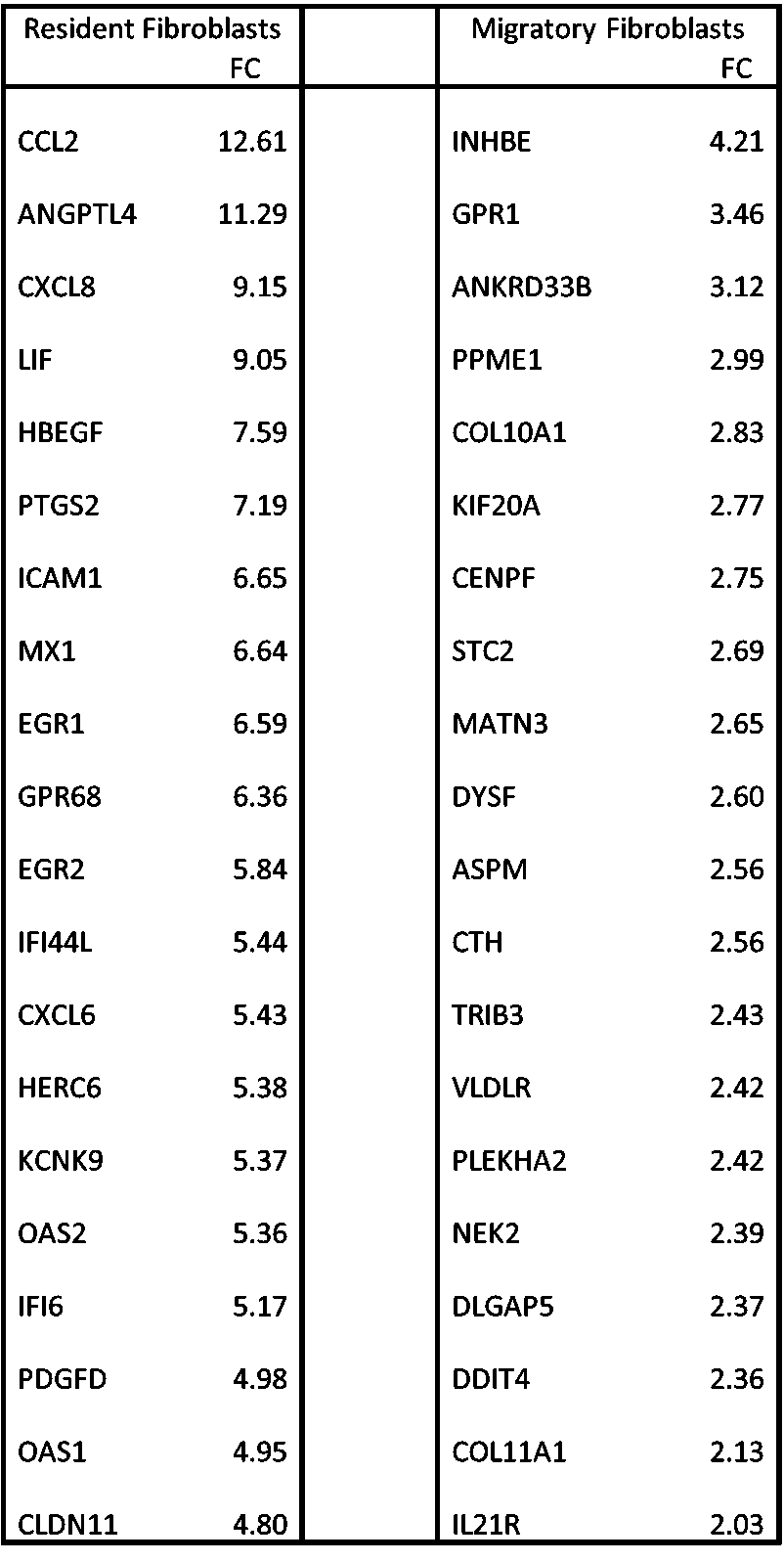
Background/Purpose: We previously described distinct migratory and resident fibroblast populations from lesional skin biopsies in SSc patients. Both populations are profibrotic compared with those from healthy control (HC) skin, however migratory SSc fibroblasts display increased expression of αSMA, greater gel contraction, and faster migration in the scratch-wound assay. The two SSc populations differ by bulk RNAseq expression, with resident fibroblasts overexpressing CCL2, CXCL8 and ICAM1, whereas migratory fibroblasts overexpress genes such as COMP, TRIP3. Here we describe the use of scRNAseq of whole skin to define the functional differences between these two fibroblasts subsets
Methods: scRNAseq was performed on 12 SSc whole skin biopsies and 3 HC, using the 10X genomics platform. The markers identified in bulk RNAseq for each fibroblast populations were used to identify corresponding clusters in scRNAseq. Candidate gene sets to identify the resident and migratory fibroblast populations were developed, and KEGG gene set enrichment pathways (GSEA) identified.
We explored differences in relative frequency of resident and migratory fibroblasts populations in whole skin by stage and subset between ATA (anti-topoisomerase) and ARA (anti-RNA pol III), as well as late and early-stage disease.
Finally, we utilised CellDive® multiplex imaging to localize the two fibroblast populations within the dermis of skin biopsies
Results: Bulk RNAseq revealed 739 genes significantly overexpressed genes in the migratory fibroblast population, whereas 745 genes were significantly upregulated in the resident fibroblasts (Table 1).
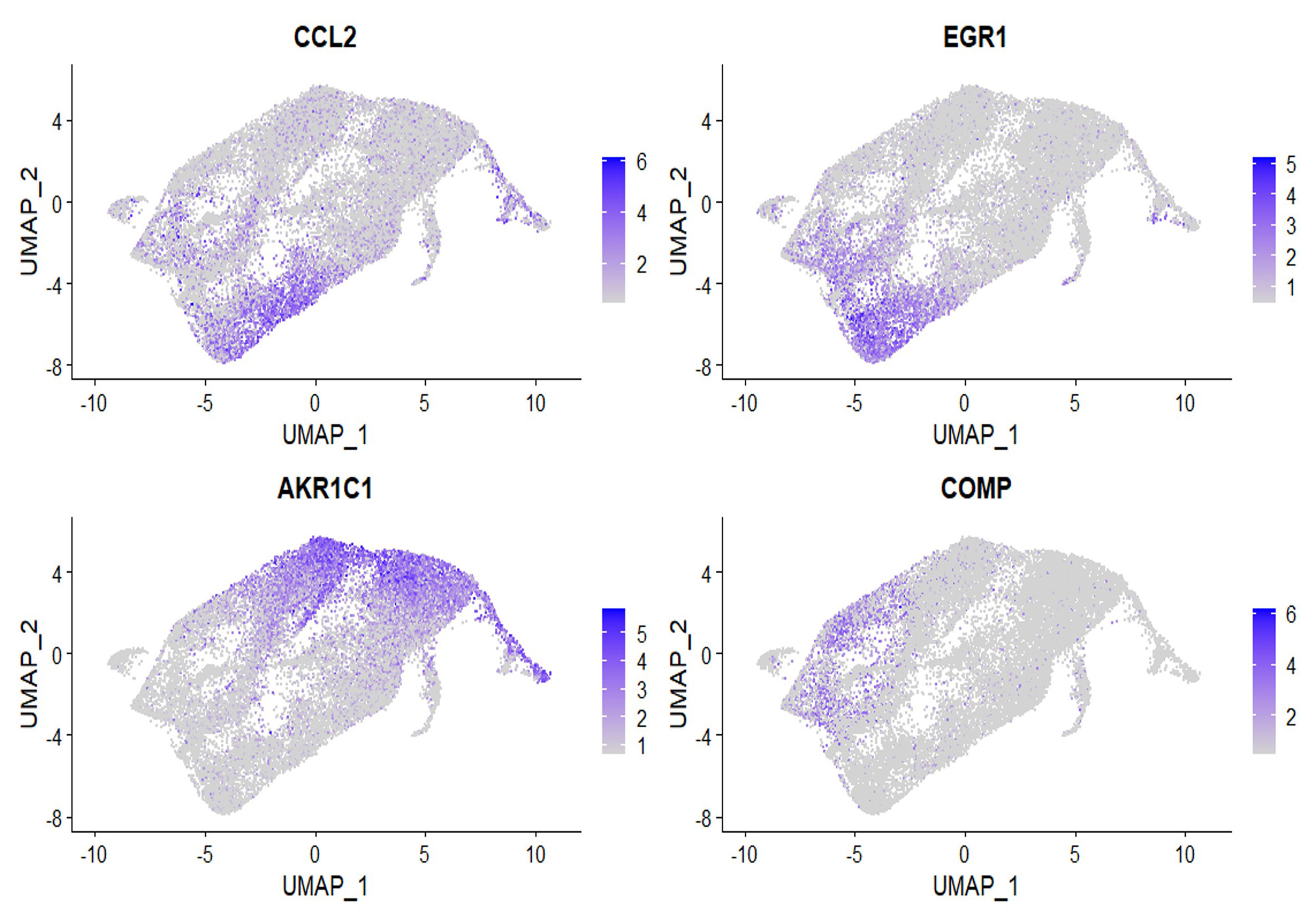
10 clusters of fibroblasts were identified on scRNAseq. Cluster 0 and 4 showed gene expression similar to migratory fibroblasts, whereas the resident fibroblasts gene expressions paralleled clusters 3 and 6 (Figure 1).
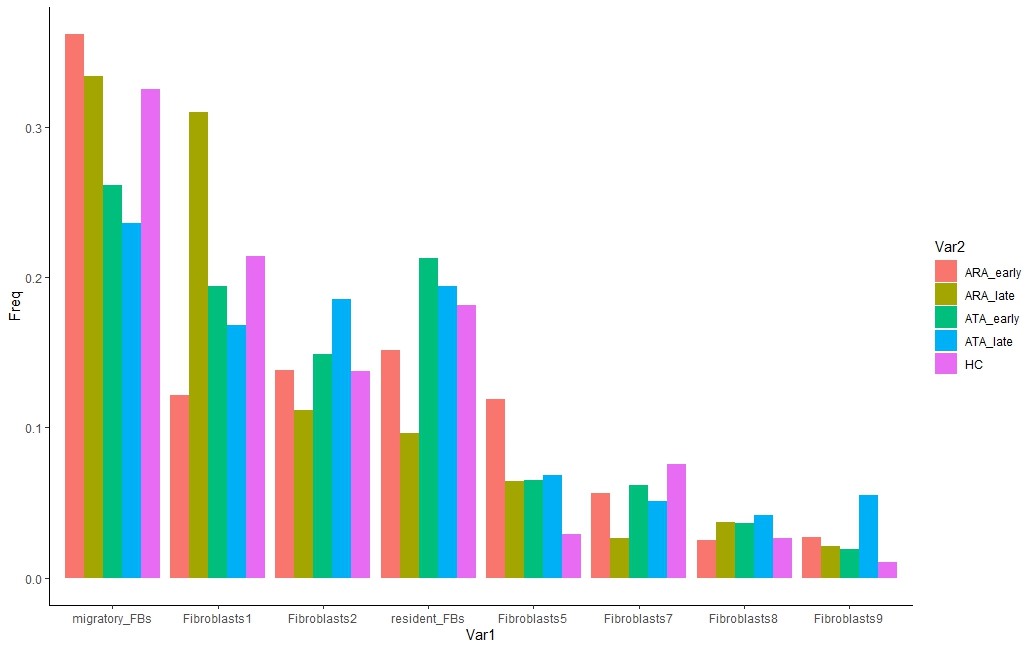
Interrogating the scRNAseq clusters, revealed the migratory fibroblasts are the most abundant cell population. Subgrouping by autoantibody identified migratory fibroblasts as most abundant in the ATA subgroup, whereas resident fibroblasts are most abundant in the ARA subgroup, suggesting autoantibody status tracks distinct fibroblast subsets (Figure 2).
GSEA of KEGG pathways confirmed concordance between migratory and resident fibroblast clusters in bulk and scRNAseq analysis. Resident fibroblasts showed upregulation of complement and coagulation genes, and JAK/STAT signalling pathway. There was upregulation of glycolysis gluconeogenesis, and PPAR signalling in migratory fibroblasts.
Using CellDive® multiplexed technology, and markers of resident and migratory fibroblasts, the fibroblast populations localise to different areas of the dermis, with migratory fibroblasts being more perivascular, and resident being scattered throughout the dermis
Conclusion: We extended our understanding of two fibroblast populations in SS and describe a molecular basis for their functional differences and location within the dermis. Understanding their different functions, and appreciating their tight association with auto antibody subset, begins to explain the differences in clinical phenotype and response to therapeutics linked to disease stage and subset of SSc.
To cite this abstract in AMA style:
Clark K, Xu S, Ong V, Buckley C, Denton C. Understanding Distinct Resident and Migratory Fibroblast Populations in Systemic Sclerosis Skin Through Single-cell RNAseq and Immunohistochemistry [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/understanding-distinct-resident-and-migratory-fibroblast-populations-in-systemic-sclerosis-skin-through-single-cell-rnaseq-and-immunohistochemistry/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/understanding-distinct-resident-and-migratory-fibroblast-populations-in-systemic-sclerosis-skin-through-single-cell-rnaseq-and-immunohistochemistry/