Session Information
Date: Monday, November 18, 2024
Title: Muscle Biology, Myositis & Myopathies – Basic & Clinical Science Poster III
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Inclusion body myositis (IBM) is a progressive muscle disease for which there is no effective treatment. Mitochondrial dysregulation is a pathologic hallmark of IBM. Pioglitazone is a potent PPARγ agonist that enhances mitochondrial biogenesis in skeletal muscle and improves mitochondrial function.
Methods: We conducted an open label phase 1 clinical trial of pioglitazone in IBM. Patients with IBM were observed for 16 weeks off therapy (lead-in period), and then received 32 weeks of pioglitazone 45 mg daily (intervention period). Serial muscle biopsies were obtained at baseline, after the lead-in period (week 16), and after 16 weeks of pioglitazone (week 32). RNA sequencing and untargeted metabolomics using mass spectrometry were performed on all muscle tissue. Metabolomics was also performed on 5 control muscle and 10 control sera. The primary endpoint was the change in PPARγ gene expression in muscle with pioglitazone. Secondary endpoints included transcriptomic and metabolic changes with therapy, and changes in clinical outcomes including the IBM-FRS, m-TUG, 6 minute walk, Fi-2, hand-grip strength, knee extensor dynamometry and creatine kinase (CK) level.
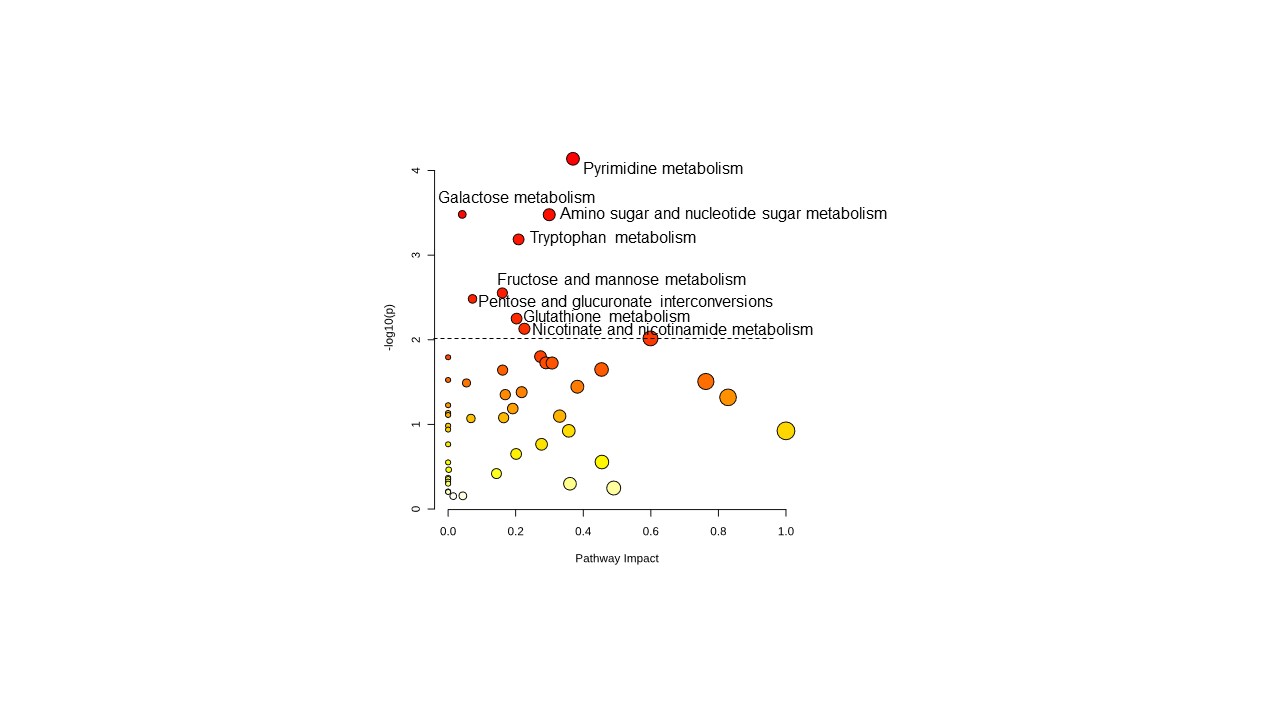
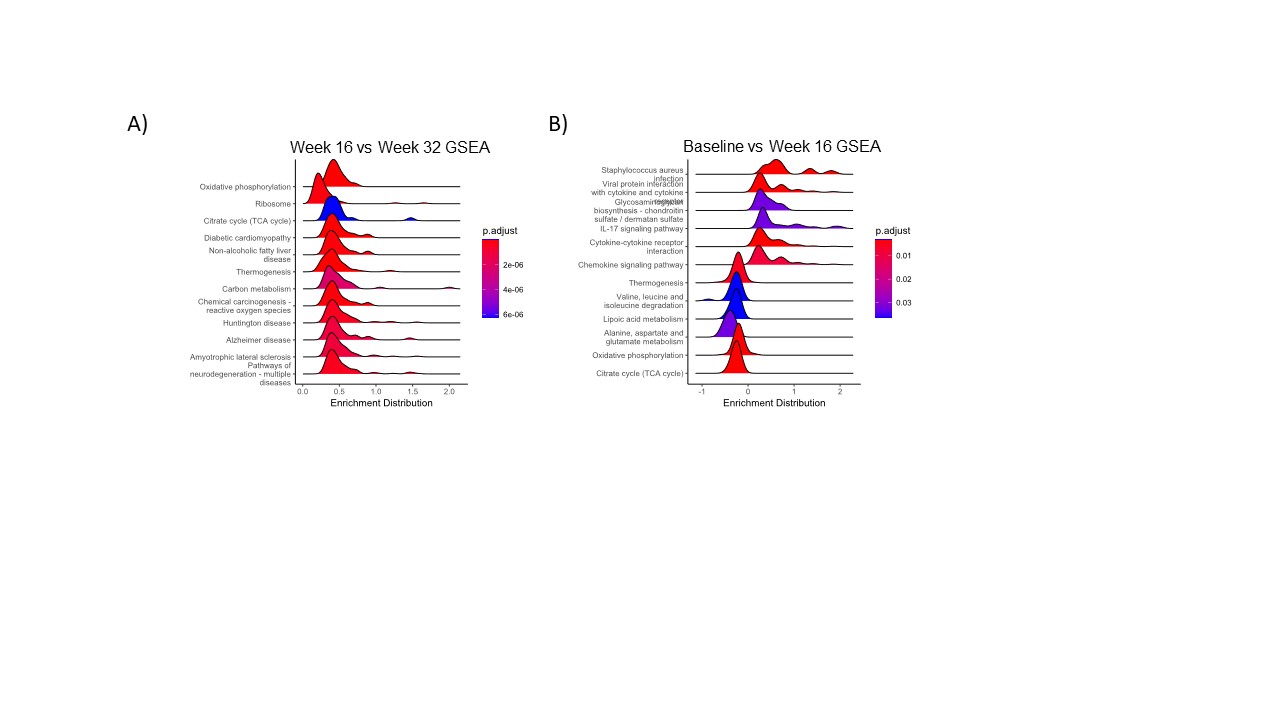
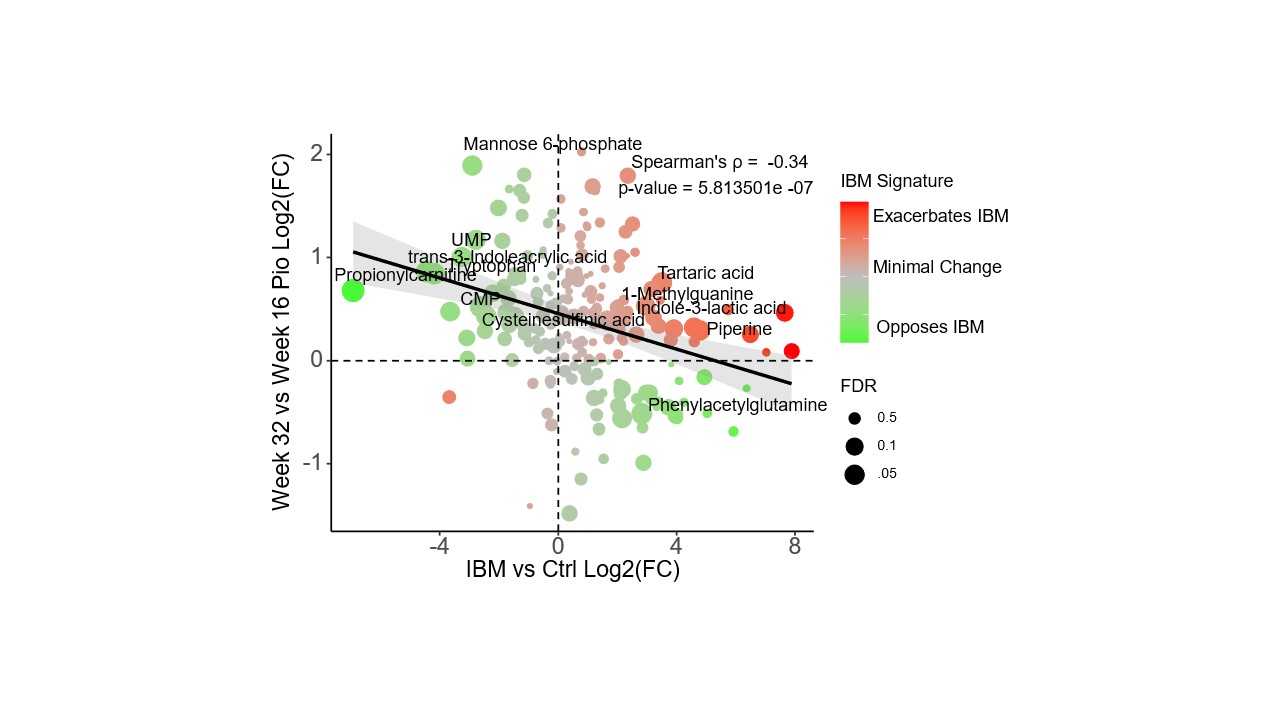
Results: 13 patients completed the lead-in phase and crossed over to receive pioglitazone. Untargeted metabolomics on baseline muscle biopsies revealed differences between IBM and control muscle (Figure 1) and sera across multiple metabolic pathways that were accentuated in more advanced disease. Analysis of paired muscle biopsies showed a nonsignificant trend of increased PPARγ gene expression with pioglitazone therapy (week32-week16 Log2 fold change (FC)=1.16, p=0.07). Gene set enrichment analysis identified increased expression of genes involved in the TCA cycle, oxidative phosphorylation, and PPAR signaling after pioglitazone (Figure 2). We performed a correlation analysis to determine how pioglitazone affects the overall IBM metabolic signature. We compared the FC values of metabolites between IBM vs. healthy control (x-axis) and post- vs. pre-treatment (y-axis). We found a significant negative correlation (Spearman’s p= -0.34, p< 0.0001), indicating that pioglitazone had a moderate effect on the muscle metabolome, shifting it towards the profile of healthy controls and away from the metabolic signature of IBM (Figure 3). The effect of pioglitazone was heterogeneous, with a positive metabolic response observed in 5/13 patients who tended to have less advanced disease. There was no measurable change in clinical outcome measures across all patients. In an exploratory analysis, we used a mixed effect model to determine the effect of metabolic response (rho) on clinical outcome. We found a significant rho x time effect for both the IBM-FRS and m-TUG, suggesting that a favorable metabolic shift away from the IBM signature was associated with a less severe decline in the IBM-FRS and a slower increase in m-TUG over time.
Conclusion: IBM is associated with metabolic dysregulation. Pioglitazone partially ameliorates some of the metabolic hallmarks of IBM, and a metabolic response in muscle may be associated with a favorable clinical trajectory. Pioglitazone warrants further investigation in IBM.
To cite this abstract in AMA style:
Adler B, Bene M, Zhang C, Paik J, Mecoli C, Lloyd T, Tiniakou E, Darrah E, Christopher-Stine L, Mears A, McGowan M, Pagkatipunan R, Le A, Albayda J. The Effects of Pioglitazone on Metabolic Dysregulation in Inclusion Body Myositis: An Open-Label Pilot Study [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/the-effects-of-pioglitazone-on-metabolic-dysregulation-in-inclusion-body-myositis-an-open-label-pilot-study/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/the-effects-of-pioglitazone-on-metabolic-dysregulation-in-inclusion-body-myositis-an-open-label-pilot-study/