Session Information
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Multiple compositional shifts of gut microbiota have been identified within the rheumatoid arthritis (RA) disease continuum, encompassing both established RA and at-risk individuals (including those with pre-clinical RA). Subsequently, a variety of gut bacteria have been implicated as a potential impetus in the development of RA, none more so than Prevotella copri. Using a comprehensively phenotyped prospective cohort of individuals at-risk of RA this work investigates whether P. copri overabundance is ubiquitous within the pre-clinical phases of RA, establishes microbiota associations with traditional RA risk factors and hypothesizes a timeline for microbial action within RA development.
Methods: Individuals at-risk of RA were defined by the presence of anti-cyclic citrullinated protein (anti-CCP) antibodies and new musculoskeletal symptoms without clinical synovitis. Individuals were sampled at baseline (n =124 participants, 30 of whom progressed to RA), and longitudinally (n = 19 participants, 5 RA progressors) at 5 time points over 15 months. At-risk individuals were compared to healthy controls (n = 22) and to individuals with new onset RA (n = 7). Taxonomic alterations of the gut microbiome were investigated using 16S rRNA amplicon sequencing, at both strain and genus level and confirmed using shotgun metagenomic DNA sequencing.
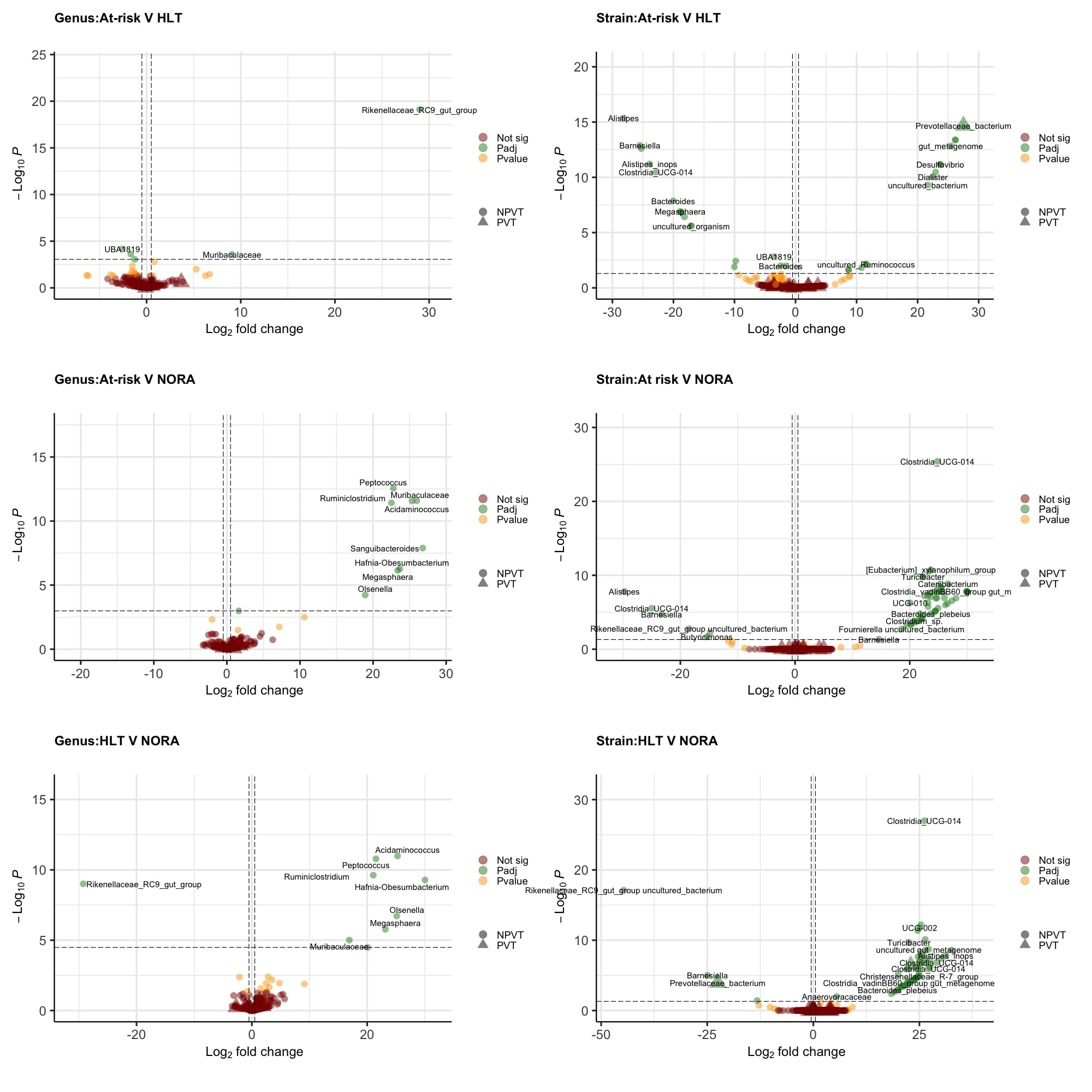
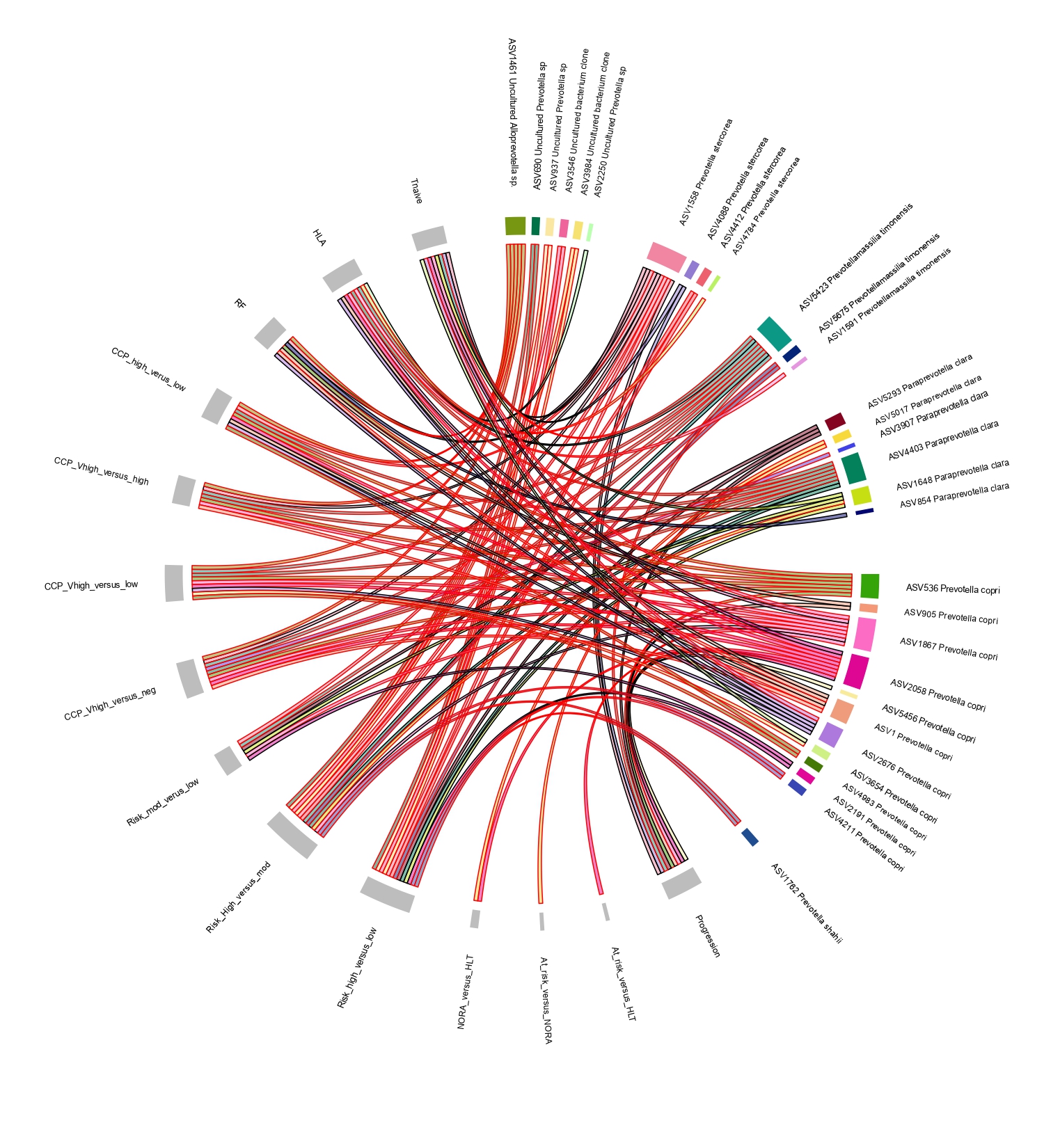
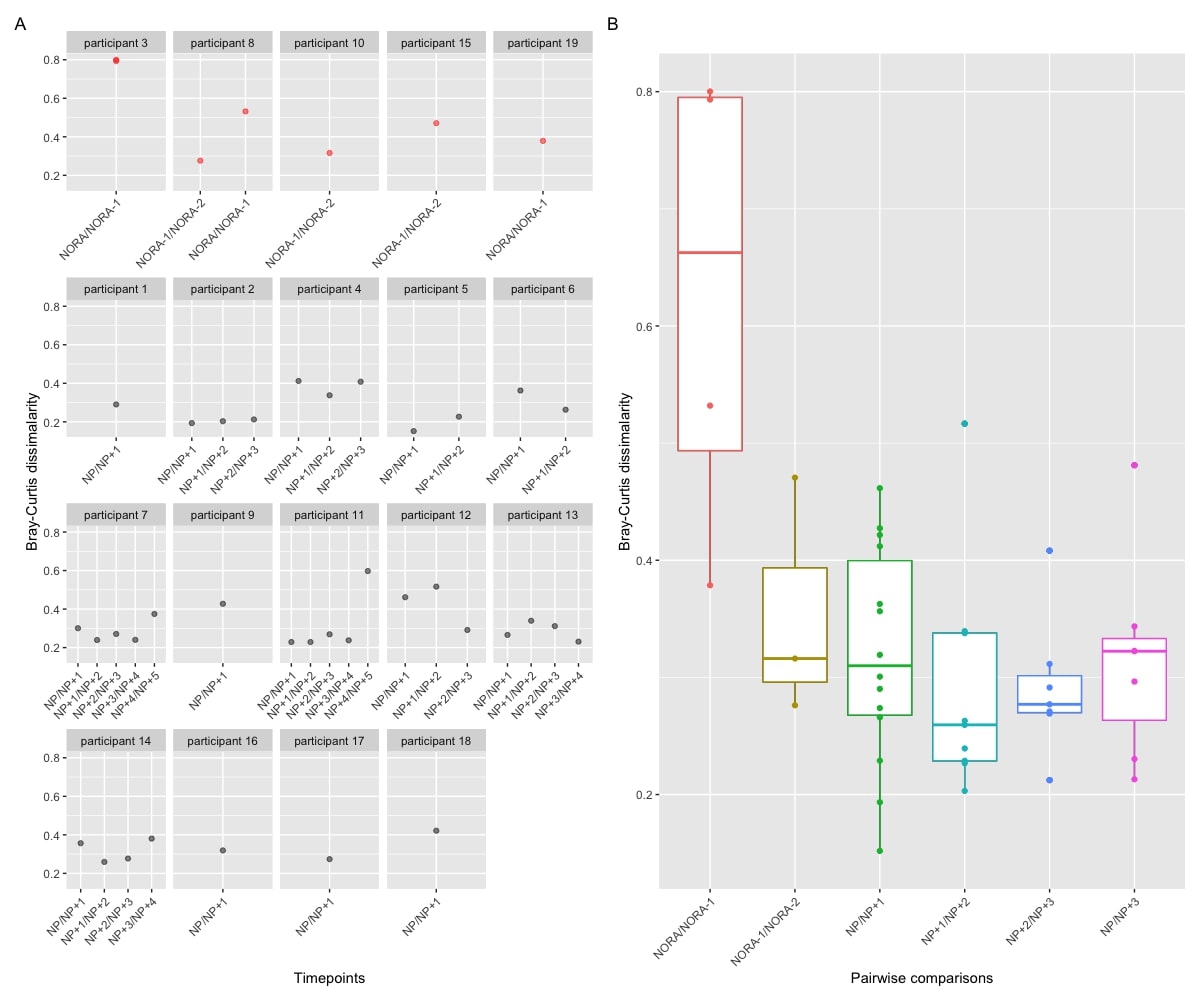
Results: RA progression at genus level was associated with modest changes in the gut microbiome, mainly affecting the Firmicutes and Proteobacteria phyla. A single Prevotellaceae strain was enriched in at-risk individuals compared to healthy controls (fig 1). P. copri strains demonstrate overall increased abundance with increasing anti-CCP levels and risk of progression (score based on symptomatology, HLA status, anti-CCP level and ultrasound findings), but display strain specific fluctuations with respect to early immune dysregulation, intestinal permeability, genetic risk and rheumatoid factor positivity (fig 2). Time series analysis suggests RA progression involves the development of an unstable low diversity microbiome that is characterised by the accumulation of RA associated pathobionts within the 10 months preceding RA (fig 3), which includes, but is not limited to P. copri. Functional analysis shows the NORA gut microbiome has increased abundance of pathways associated with amino acid metabolism. Healthy controls had decreased ornithine production, a precursor for citrulline.
Conclusion: These results confirm and expand the bacterial associations linked to RA progression, which involves multiple taxa at both genus and strain level and is associated with the underlying risk profile of an individual. As with previous literature, an overabundance of Prevotellaceae was identified in those at-risk of RA compared to healthy controls. Strain specific fluctuations appears to be the hallmark of Prevotellaceae associations within the RA at-risk phase. Future work will investigate how niche bacterial specialisation to the underlying risk profile may subsequently affect the abundance threshold required for a bacterial impetus in RA related autoimmunity.
To cite this abstract in AMA style:
Rooney C, Jeffery I, Mankia K, Wilcox M, Emery P. The Dynamics of the Gut Microbiome in Rheumatoid Arthritis Susceptibility: A Cross-Sectional and Longitudinal Observational Study [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/the-dynamics-of-the-gut-microbiome-in-rheumatoid-arthritis-susceptibility-a-cross-sectional-and-longitudinal-observational-study/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/the-dynamics-of-the-gut-microbiome-in-rheumatoid-arthritis-susceptibility-a-cross-sectional-and-longitudinal-observational-study/