Session Information
Date: Saturday, November 16, 2024
Title: Abstracts: Osteoarthritis – Novel Insights from Observational Studies
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Knee osteoarthritis (OA) is a heterogeneous disease characterized by a variety of clinical and molecular phenotypes. However, we do not yet have robust biomarkers to distinguish/predict these phenotypes. We previously published analyses of baseline peripheral blood cell DNA methylation patterns to predict future radiographic and pain progression in patients with early symptomatic knee OA. In the current study, we expand this previous analysis to include the prediction of future incident radiographic knee OA using baseline peripheral blood DNA methylation data from healthy individuals in the Osteoarthritis Initiative (OAI) cohort who experienced incident radiographic OA development during the study.
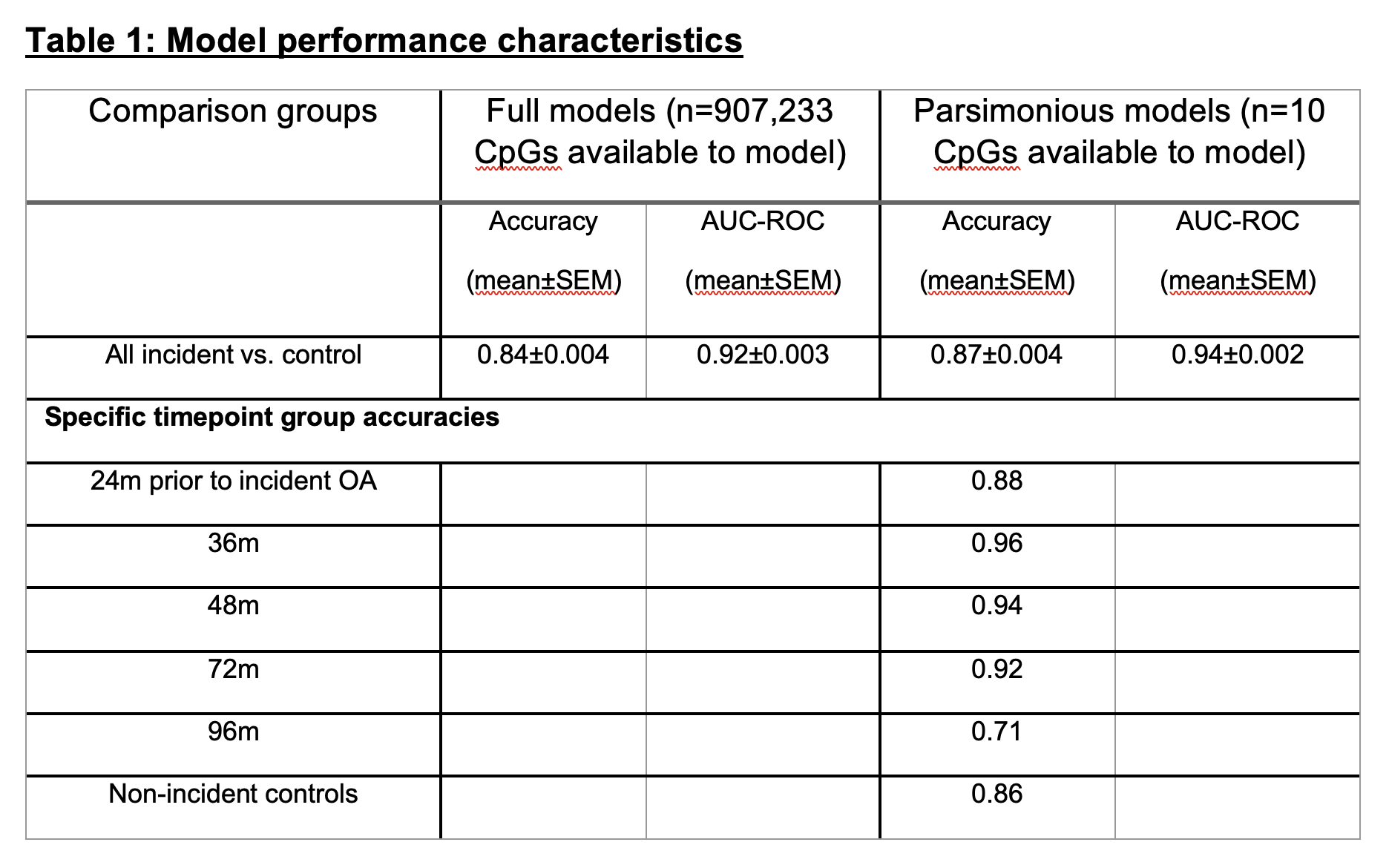
Methods: Baseline-blood-draw buffy coat DNA was obtained from the OAI (n=236 future OA cases, n=236 controls). Incident OA patients were defined as baseline K/L radiographic grade 0-1 with follow-up K/L grade≥2 between 24 and 96 months after baseline (42 at 24m, 47 at 36m, 35 at 48m, 63 at 72m, and 49 at 96m. Controls were matched by age, BMI, ethnicity, and sex. DNA (500ng) was bisulfite treated and methylation quantified using Illumina EPICv2 arrays. Generalized logistic models (GLMs) including automated feature selection and reduction were developed to classify samples using 40 cycles of training (70%) and testing divisions (30%). Parsimonious models were applied to training and test sets by including DNA methylation sites and covariates that were included in at least 20 of 40 development cycles. Previous parsimonious CpG sites identified in progression studies of established OA were also used to develop incident models.
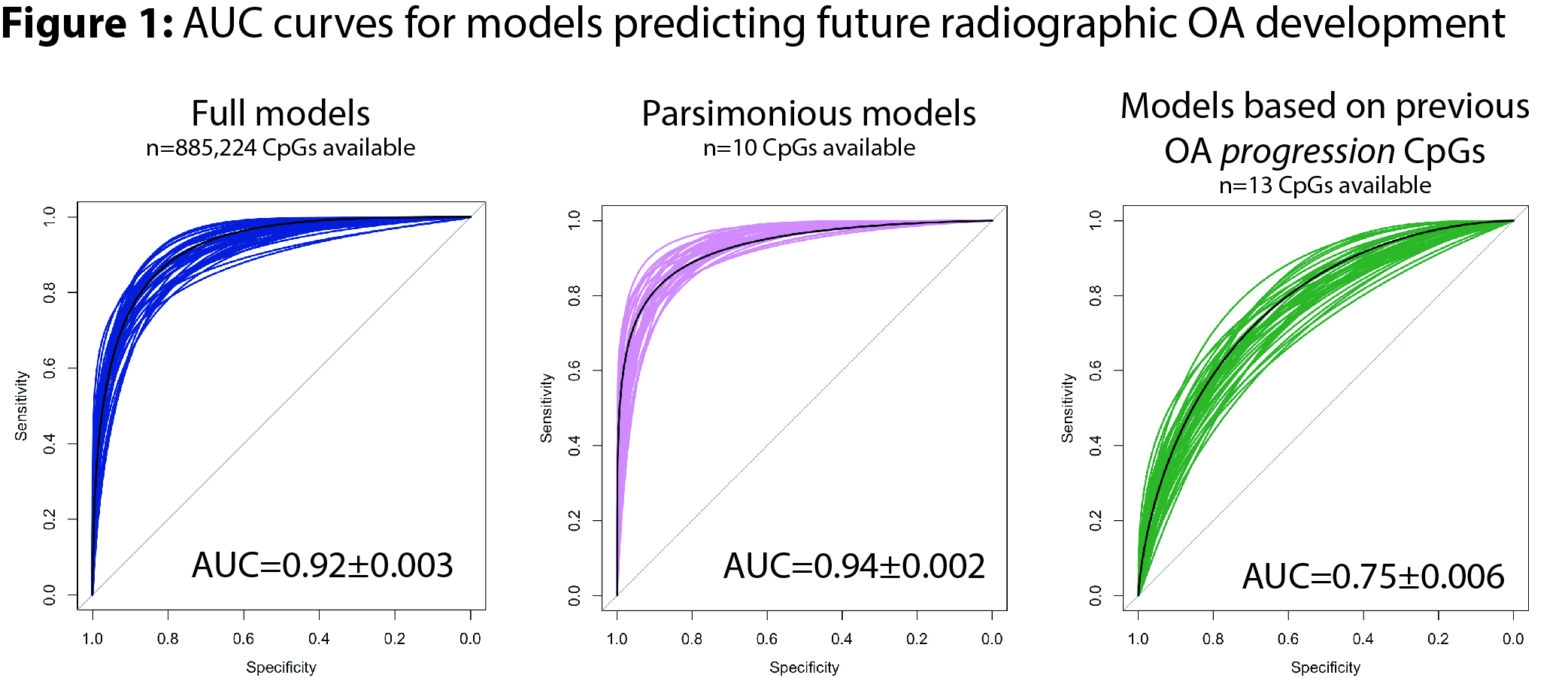
Results: Baseline buffy coat DNA methylation patterns accurately differentiated future incident OA cases from controls (Table 1, Figure 1). Including all time points, full models (n=885,224 CpG sites available) demonstrated AUC=0.92±0.003 (mean±SEM), accuracy on unseen test data=0.84±0.004. Parsimonious models (n=10 CpG sites available) showed improved discriminatory capability with AUC=0.94±0.002, accuracy=0.87±0.004, P=6E-7 vs. full models). Model accuracy was highest at 36m prior to OA onset (accuracy 0.96), whereas 96m model accuracy was the lowest (0.71). Demographic covariates were not predictive of future incident OA. CpG sites identified in our previous radiographic progression modeling (in established OA patients) were also somewhat predictive of future OA development, albeit at a lower accuracy (AUC=0.75±0.006, accuracy=0.69±0.005).
Conclusion: Herein, we generated peripheral blood DNA methylation data on baseline blood draws from OAI patients who went on to develop incident radiographic knee OA within 24-96 months and compared this to control patients without incident OA. Models demonstrated high accuracy at all time points, peaking at 36m prior to OA development. Future work should focus on confirming these findings in additional cohorts as well as evaluating differential DNA methylation of individual peripheral blood immune cell populations in individuals with incident knee OA.
To cite this abstract in AMA style:
Miranda C, Hannebutt N, dyson G, Barrett M, Szymczak A, Jeffries M. The Development of Future Radiographic Knee Osteoarthritis Can Be Predicted by Peripheral Blood Epigenetic Models [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/the-development-of-future-radiographic-knee-osteoarthritis-can-be-predicted-by-peripheral-blood-epigenetic-models/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/the-development-of-future-radiographic-knee-osteoarthritis-can-be-predicted-by-peripheral-blood-epigenetic-models/