Session Information
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Skin inflammation in juvenile dermatomyositis (JDM) can signal disease onset or flare and prevent complete disease remission. The study of cutaneous expression signatures holds the potential to reveal novel mechanisms underlying disease heterogeneity and organ-specific inflammation. The objectives in this study were to 1) analyze skin tape stripping (TS) transcriptional profiles to identify JDM disease endotypes and 2) compare skin and blood expression signatures.
Methods: We performed TS on non-lesional +/- lesional skin in a JDM cohort (n=28, n=16 with longitudinal sampling, n=77 TS samples). All JDM patients met 2017 EULAR/ACR classification criteria and had standardized clinical disease activity assessments. Paired blood was collected in PAXgene tubes. mRNA was isolated from TS and blood followed by RNAseq. Unsupervised hierarchical clustering was performed to determine molecular JDM subgroups. Differentially expressed genes (DEGs; q-value< 0.01 and absolute log2 fold-change ≥ 1) between subgroups were extracted using TIGR-MeV. Literature-based pathways were explored using Ingenuity Pathway Analysis (IPA). Pathway-based expression scores in skin and blood were calculated (PMID ref: 16947629) and compared between subgroups (t-test; p-value< 0.05). Cell type enrichment analysis was performed using xCell.
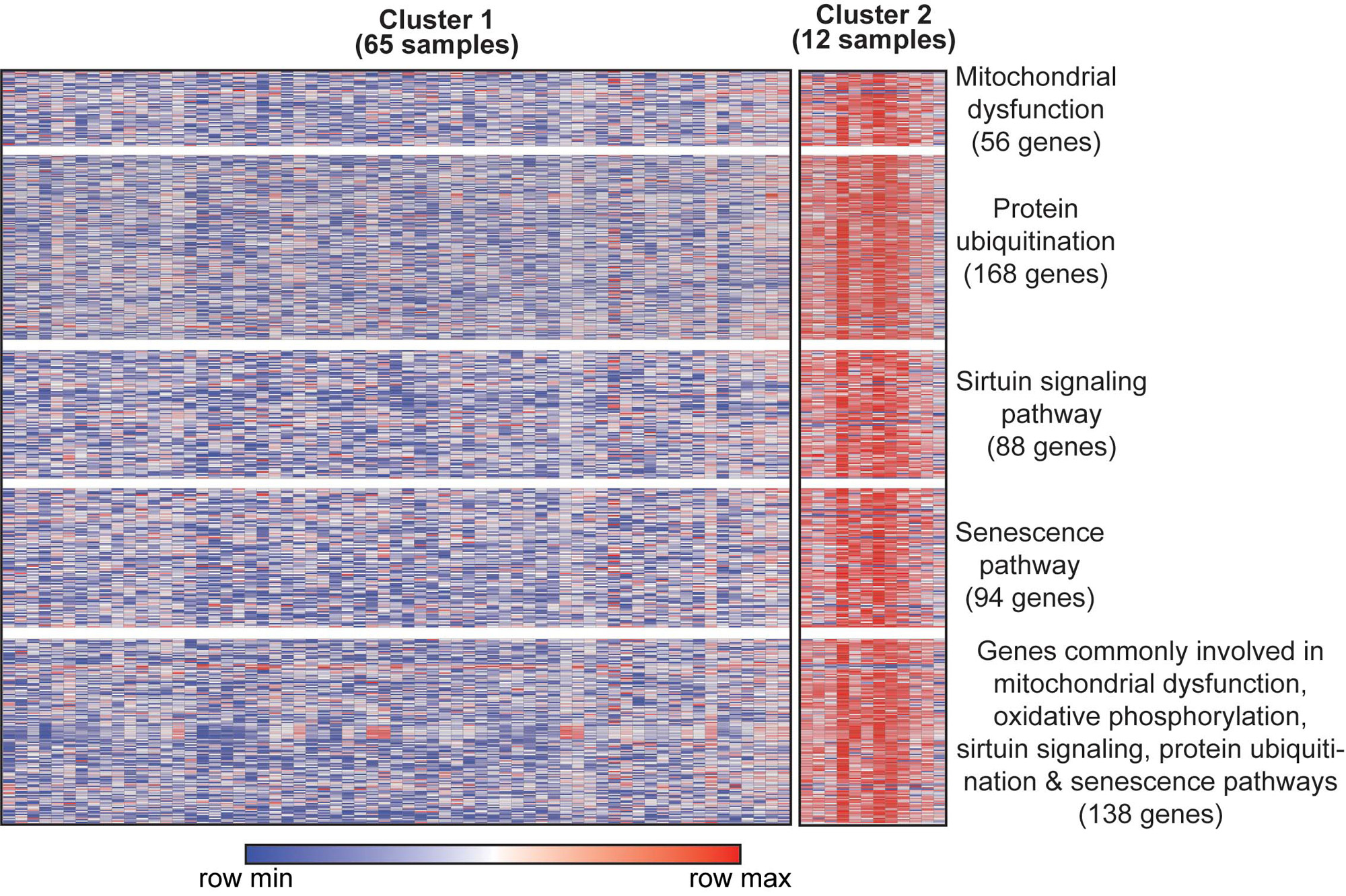
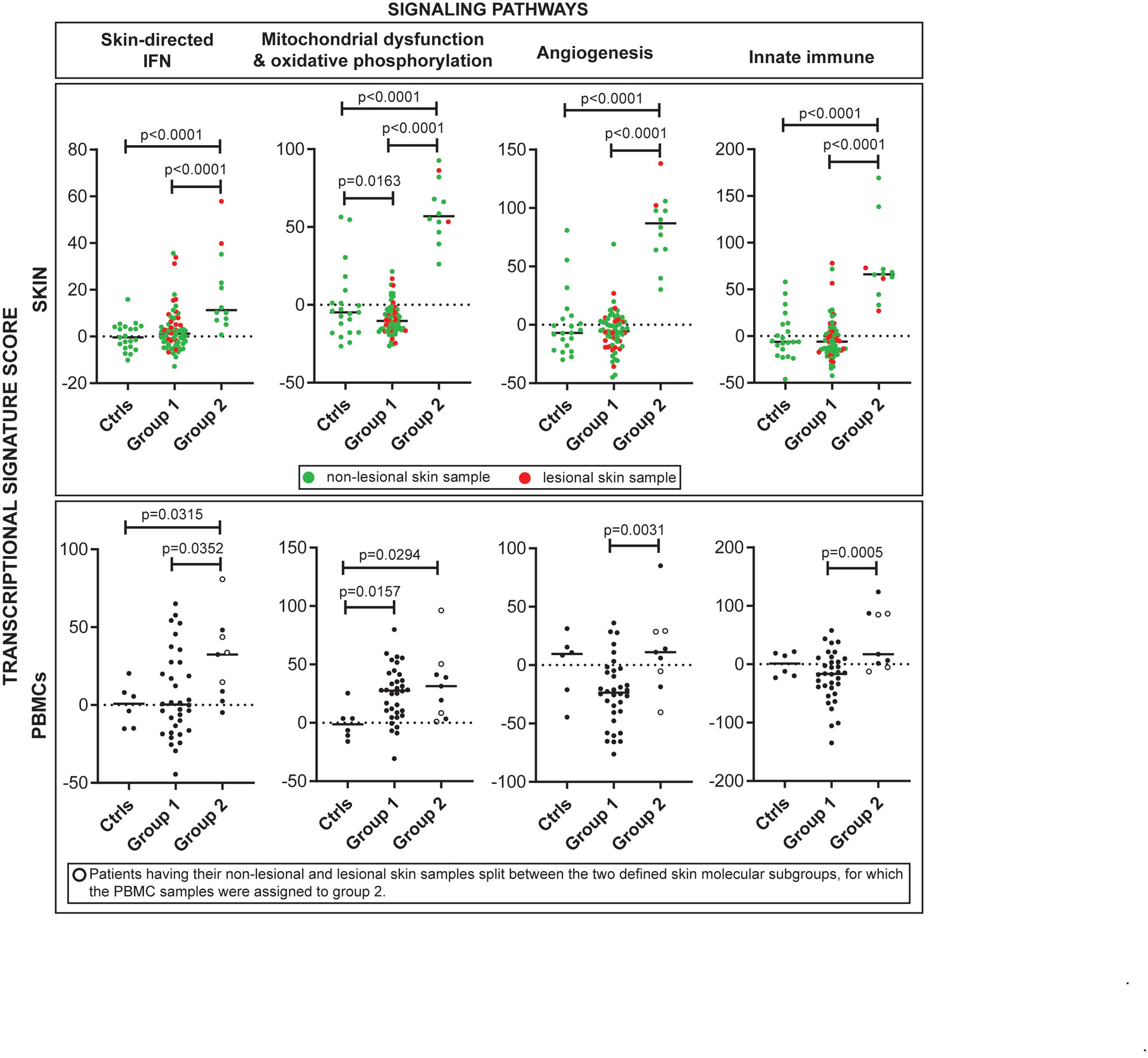
Results: We identified two JDM subgroups with distinct TS expression profiles (corresponding to 65 samples (28 patients) in cluster 1 and 12 samples (8 patients) in cluster 2). The two subgroups did not separate by lesional/non-lesional skin, disease duration or activity. However, subgroup 2 represented patients with a higher frequency of chronic disease and more likely to be on steroids. The 6,773 DEGs distinguishing the two subgroups represented pathways involving mitochondrial dysfunction, sirtuin signaling, oxidative phosphorylation, protein ubiquitination and senescence (p-value< 0.0001) (Figure 1). NFE2L2, a transcription factor involved in response to oxidative stress and innate immune signaling, was the top upstream regulator activated in subgroup 2 (IPA Z-score = 10.74, enrichment p-value = 6.1 E-14). Subgroup 2 demonstrated higher skin-directed interferon, mitochondrial/oxidative phosphorylation dysfunction, angiogenesis and innate immune scores in skin compared to subgroup 1 and controls (Figure 2, top panel). Blood as compared to skin-derived pathway scores did not as effectively highlight biological differences in subgroup 2 (Figure 2, lower panel). Subgroup 2 also differed from subgroup 1 in potential immune cell populations present in skin, notably with higher xCell scores from CD4 T-cells in subgroup 2 and from B-cells in subgroup 1.
Conclusion: We identified a unique JDM subgroup based on TS expression signatures, with upregulation of genes involved in mitochondrial dysfunction, sirtuin signaling and protein ubiquitination. Interestingly, TS JDM subgroups were distinguished by metabolic in addition to traditional inflammatory pathways. TS performed better than blood in differentiating possible mechanisms underlying JDM disease endotypes. Skin-specific transcriptomic signatures may be able to lend insight into precision medicine care in JDM.
To cite this abstract in AMA style:
Turnier J, Berthier C, Vandenbergen S, Goudsmit C, McClune M, Gudjonsson J, Tsoi L, Kahlenberg J. Tape Stripping Expression Signatures Identify Biologically Unique Juvenile Dermatomyositis Patient Subgroup Characterized by Increased Mitochondrial Dysfunction [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/tape-stripping-expression-signatures-identify-biologically-unique-juvenile-dermatomyositis-patient-subgroup-characterized-by-increased-mitochondrial-dysfunction/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/tape-stripping-expression-signatures-identify-biologically-unique-juvenile-dermatomyositis-patient-subgroup-characterized-by-increased-mitochondrial-dysfunction/