Session Information
Date: Sunday, October 26, 2025
Title: Abstracts: Rheumatoid Arthritis – Etiology and Pathogenesis (0795–0800)
Session Type: Abstract Session
Session Time: 1:30PM-1:45PM
Background/Purpose: The synovial lining is crucial for joint homeostasis, forming a barrier and producing lubricants through specialized fibroblasts. These fibroblasts differ from sublining fibroblasts, which are involved in inflammation and matrix remodeling. While vascular Notch signaling helps define vascular fibroblast positional identity (Wei et al. 2020), the factors shaping synovial lining fibroblast identity are unclear. Using spatial transcriptomics on synovial biopsies from RA patients in remission, we identify a density-dependent mechanism regulating lining fibroblast identity and suggest CREB5, SOX5, and FOXO1 as key transcriptional regulators.
Methods: Synovial tissue and synovial fibroblast micromasses were processed using the Xenium In Situ Gene Expression protocol (10X Genomics, CG000582 Rev D) with a custom 400-gene panel(Bhamidipati et al. 2025). For bulk RNA-seq and qRT-PCR, synovial fibroblasts were seeded at densities of 75–600 cells/mm² in 96-well plates for 3 days. For siRNA knockdown, cells were transfected with siRNA at 200K/well in 6-well plates and then re-seeded at varying densities.
Results: Spatial transcriptomics identified two fibroblast clusters in synovial tissue from RA patients in clinical remission: lining fibroblasts in dense, peripheral regions and sublining fibroblasts in less dense, central areas. A similar spatial organization was observed in synovial fibroblast micromass cultures maintained for 21 days. Extended culture led to increased accumulation of sublining fibroblasts in the central region, while lining fibroblasts remained localized to the periphery. Among the genes enriched in high-density regions of synovial tissue and fibroblast micromass cultures were the transcription factors CREB5, SOX5, and FOXO1, which exhibited spatial distribution patterns similar to canonical lining markers (PRG4, CLU, PDPN). These factors emerged as key candidates for regulating fibroblast identity. In vitro knockdown of CREB5 significantly reduced lining marker expression and increased sublining markers in a density-dependent manner. Similarly, knockdown of SOX5 and FOXO1 decreased lining marker expression without significantly affecting sublining marker levels.
Conclusion: In summary, our findings identify a density-responsive subset of lining fibroblasts defined by elevated CREB5, SOX5, and FOXO1 expression. We propose that increased cell density may activate as-yet-unidentified upstream signaling pathways that modulate the expression and/or activity of CREB5, FOXO1, and SOX5. Acting individually or synergistically, these transcription factors appear to mediate a cell density–dependent phenotypic transition from sublining to lining fibroblast states, thereby contributing to fibroblast identity.
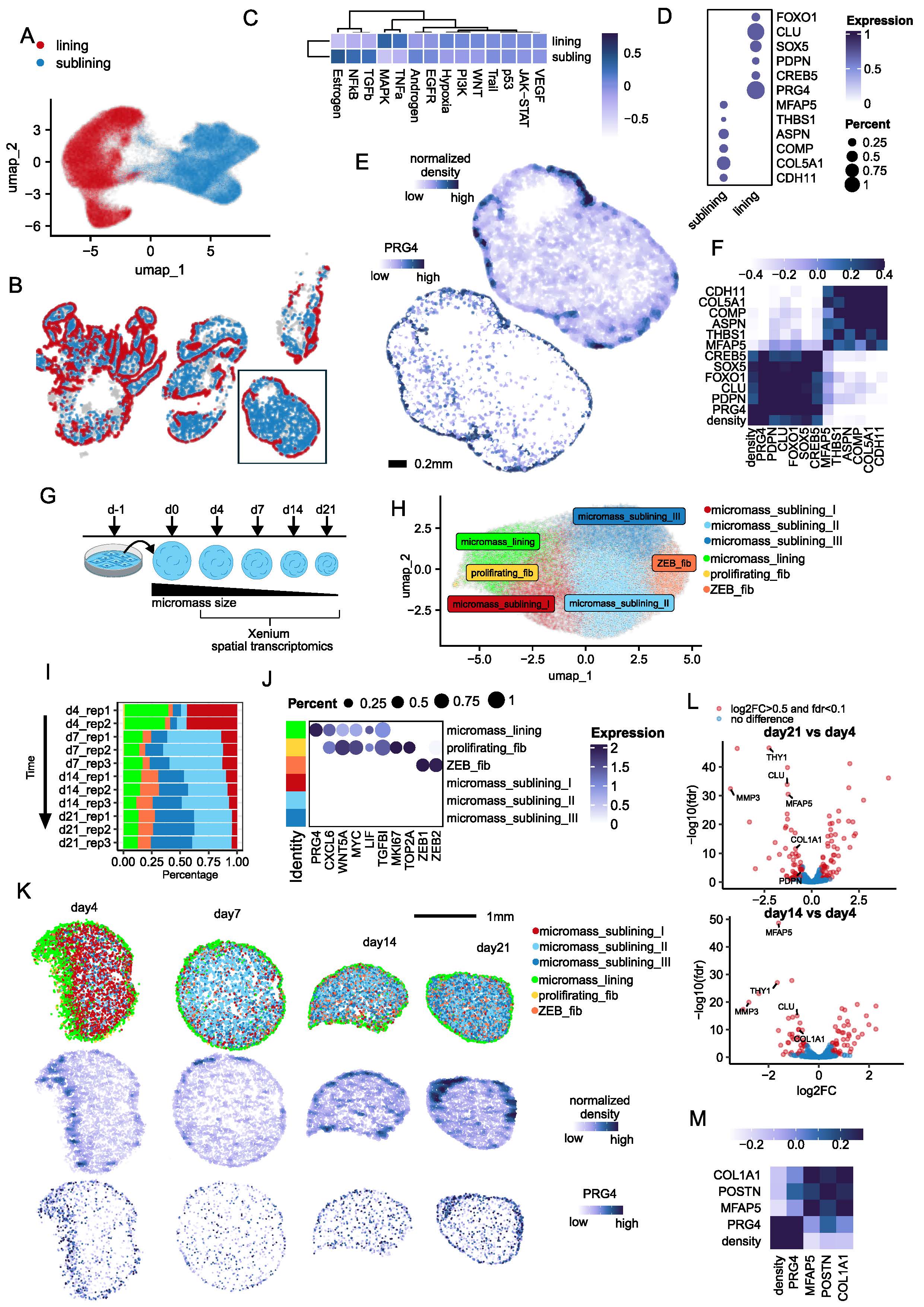 Figure 1. Characterization of fibroblast populations in RA synovial tissue and synovial fibroblast micromass culture system
Figure 1. Characterization of fibroblast populations in RA synovial tissue and synovial fibroblast micromass culture system
.jpg) Figure 2. Bulk RNA-seq analysis of CREB5 KD (A-F) and qRT-PCR analysis of SOX5 and FOXO1 KD in synovial fibroblasts
Figure 2. Bulk RNA-seq analysis of CREB5 KD (A-F) and qRT-PCR analysis of SOX5 and FOXO1 KD in synovial fibroblasts
To cite this abstract in AMA style:
Presti S, Anufrieva K, Gao C, Prell S, Blazar P, Lange J, Jones M, Wechalekar M, Brenner M, Korsunsky I, Kazerounian S, Wei K. Spatial transcriptomics identifies density-sensing fibroblasts in synovial lining [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/spatial-transcriptomics-identifies-density-sensing-fibroblasts-in-synovial-lining/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/spatial-transcriptomics-identifies-density-sensing-fibroblasts-in-synovial-lining/
