Session Information
Date: Tuesday, October 28, 2025
Title: (2227–2264) Rheumatoid Arthritis – Diagnosis, Manifestations, and Outcomes Poster III
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Autoimmune diseases affect more than 23.5 million Americans involving nearly every organ system of the human body. To identify shared, unique, and novel pathways underlying the pathogenesis of common autoimmune diseases, The Accelerating Medicines Partnership: Autoimmune and Immune-Mediated Diseases Network (AMP AIM) was launched to investigate Rheumatoid Arthritis (RA), Systemic Lupus Erythematosus (SLE), Psoriatic Disease (PsD), and Sjogren’s Disease (SD) using single-cell and spatial technologies.
Methods: The Disease Teams, Technology Cores, System Biology Groups, Tissue Research Center, and Research Management Units collaborated on pilot projects to evaluate spatial and single-cell technologies for their feasibility, sensitivity, and scalability. These assessments aimed to inform the selection of omics approaches that would provide the greatest utility and reproducibility across the network’s diverse research settings.
Results: Following two years of focused investigation, the AMP AIM network developed a standard operating procedure which enables a multi-institutional, spatial multi-omics workflow of deep phenotyping of study participants. This protocol leverages centrally collected, FFPE-preserved target tissues (e.g., synovium, skin, salivary gland, and kidney) and integrates spatial transcriptomics (ST) using Xenium (10x Genomics), histological evaluation via H&E staining, and protein-level spatial profiling with Imaging Mass Cytometry (IMC) on the Hyperion platform (Standard BioTools), all on the same slide.1. Successful implementation of combined spatial transcriptomics and proteomics. With the network selecting Xenium as the preferred ST platform and post-run H&E stain, we developed a workflow which integrates Xenium and IMC from the same tissue section. This results in a unified dataset comprising ~5,000 spatially resolved transcripts, 40 protein markers, and histological context, enabling single-cell level correlation of gene and protein expression within intact tissue architecture (Fig 1).2. Defining shared immune cell states in target tissues across RA, SLE, PSD, and SD. To identify shared immune cell states across autoimmune diseases and target tissues, we implemented a computational pipeline for robust cell type and state annotation in spatial transcriptomic datasets. To evaluate the generalizability of this pipeline, we used a unified scRNA-seq reference to annotate T and NK cell states across spatially profiled tissues. This approach identified 9 distinct T cell states (Fig 2A), each exhibiting comparable transcriptional signatures across RA, SLE, PSD, and SD.
Conclusion: This integrated spatial multi-omics platform is being applied across disease-specific (e.g., RA, SLE, PSD, and SD) and cross-disease studies to uncover shared and distinct cellular programs in autoimmune pathogenesis. Through robust, high-resolution, single cell analyses within the tissue context, our approach lays the foundation for discovering common and unique mechanisms and advancing targeted therapeutics across autoimmune diseases.
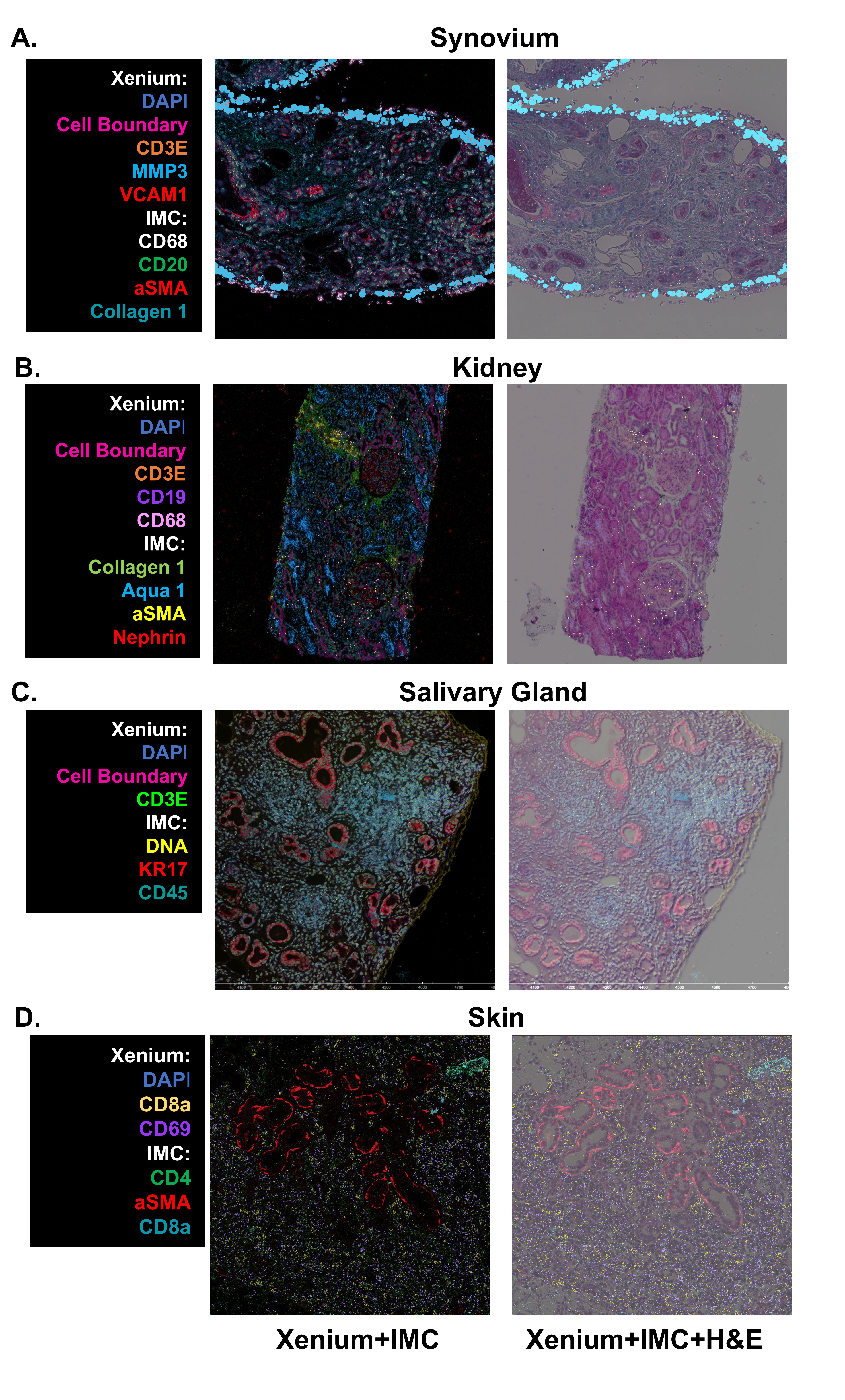 Figure 1: Spatial Characterization of Target Tissues. Representative images from all four target tissues (Synovium, Kidney, Salivary Gland, and Skin) processed and imaged on the Xenium spatial transcriptomics (10X Genomics), stained and whole slide imaged for H&E, and then stained and imaged for ~40 surface proteins via Imaging Mass Cytometry (Standard Biotools), all on the same tissue section and slide. Similar cell types (IMC) or transcripts (Xenium) are highlighted in the example projections to demonstrate agreement across technologies.
Figure 1: Spatial Characterization of Target Tissues. Representative images from all four target tissues (Synovium, Kidney, Salivary Gland, and Skin) processed and imaged on the Xenium spatial transcriptomics (10X Genomics), stained and whole slide imaged for H&E, and then stained and imaged for ~40 surface proteins via Imaging Mass Cytometry (Standard Biotools), all on the same tissue section and slide. Similar cell types (IMC) or transcripts (Xenium) are highlighted in the example projections to demonstrate agreement across technologies.
.jpg) Figure 2: Assessment of Spatial Transcriptomics Technologies demonstrates shared T-cell states across tissues. (A) Median abundance of T-cell states, shown as the proportion relative to the number of T-cells within each tissue. (B-E) Spatial localization of T-cell states within selected regions of interest. in: (B) skin, (C) salivary gland, (D) synovium, and (E) kidney tissues. T-cells are colored by their state, and non-T-cells are colored in grey.
Figure 2: Assessment of Spatial Transcriptomics Technologies demonstrates shared T-cell states across tissues. (A) Median abundance of T-cell states, shown as the proportion relative to the number of T-cells within each tissue. (B-E) Spatial localization of T-cell states within selected regions of interest. in: (B) skin, (C) salivary gland, (D) synovium, and (E) kidney tissues. T-cells are colored by their state, and non-T-cells are colored in grey.
To cite this abstract in AMA style:
Marlin C, Madhu R, Theisen E, Bradshaw L, Gao C, Eisenhaure T, Sugiarto N, Autoimmune and Immune Mediated Diseases A, Buyon J, Petri M, Rovin B, Werth V, Gravallese E, Anolik J, Moreland L, Donlin L, Ritchlin C, Scher J, Gudjonsson J, Liao W, Ogdie A, Shiboski C, Farris A, Baer A, Warner B, Clemente J, Heguy A, Brenner M, Hacohen N, Guthridge J, James J, Raychaudhuri S, Tsoi L, Zhou X, Welch J, Clark R, Korsunsky i, Lessard C, Wei K. Spatial Multi-omics Demonstrates Shared Immune States Across Autoimmune Diseases and Target Tissues in the Accelerating Medicines Partnership: Autoimmune and Immune-Mediated Diseases Network (AMP AIM) [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/spatial-multi-omics-demonstrates-shared-immune-states-across-autoimmune-diseases-and-target-tissues-in-the-accelerating-medicines-partnership-autoimmune-and-immune-mediated-diseases-network-amp-aim/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/spatial-multi-omics-demonstrates-shared-immune-states-across-autoimmune-diseases-and-target-tissues-in-the-accelerating-medicines-partnership-autoimmune-and-immune-mediated-diseases-network-amp-aim/
