Session Information
Session Type: Abstract Session
Session Time: 4:00PM-5:30PM
Background/Purpose: Spinal ligament degeneration is a chronic disease that affects the spine, ligaments, and associated bones, resulting in back pain and functional limitations in the world’s aging population. Despite the significant burden on musculoskeletal medicine, there is a lack of research on preventive strategies for ligament degeneration. This study aims to establish a comprehensive transcriptomic atlas of ligament tissues and identify effective drug targets.
Methods: Single-cell RNA sequencing was performed on six degenerative ligaments and three traumatic ligaments to decipher the heterogeneity of the tissues, followed by data analysis using various computational tools. Library preparation with 10X Genomics and sequencing on Illumina NextSeq instrument was performed. Immunohistochemistry and immunofluorescence analysis were performed to validate the molecular patterns and while in vitro models of primary ligament cells were isolated and utilized for the follow-up functional studies. Protein and mRNA levels were examined using western blot and qPCR analysis. GW5074 (an inhibitor targeting ATF3) and si-ATF3 were used to silence ATF3 in ligament cells, and lentiviral transfection was conducted to overexpress ATF3. In addition, ligament cells were stimulated with recombinant SPP1 to evaluate its impact.
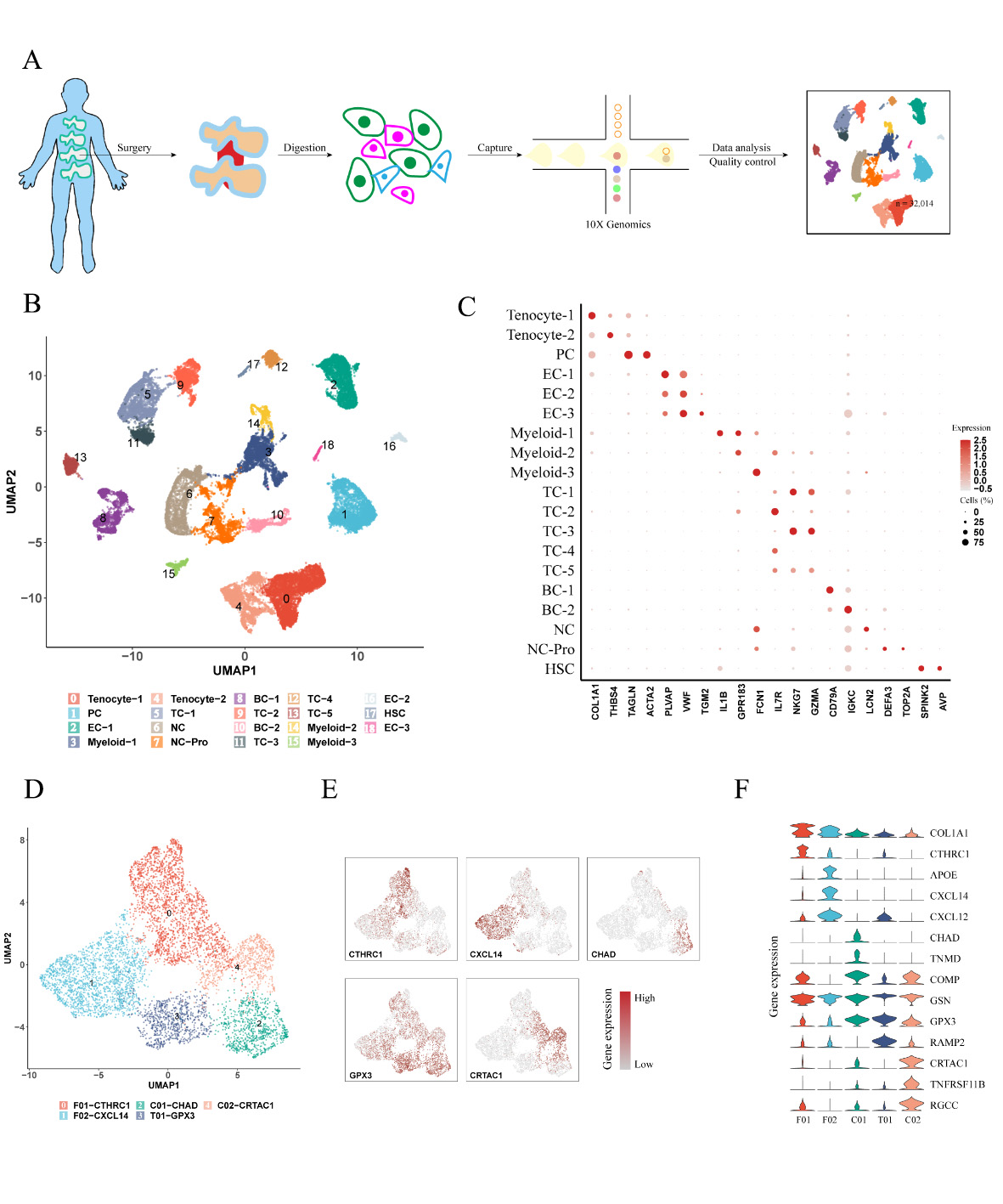
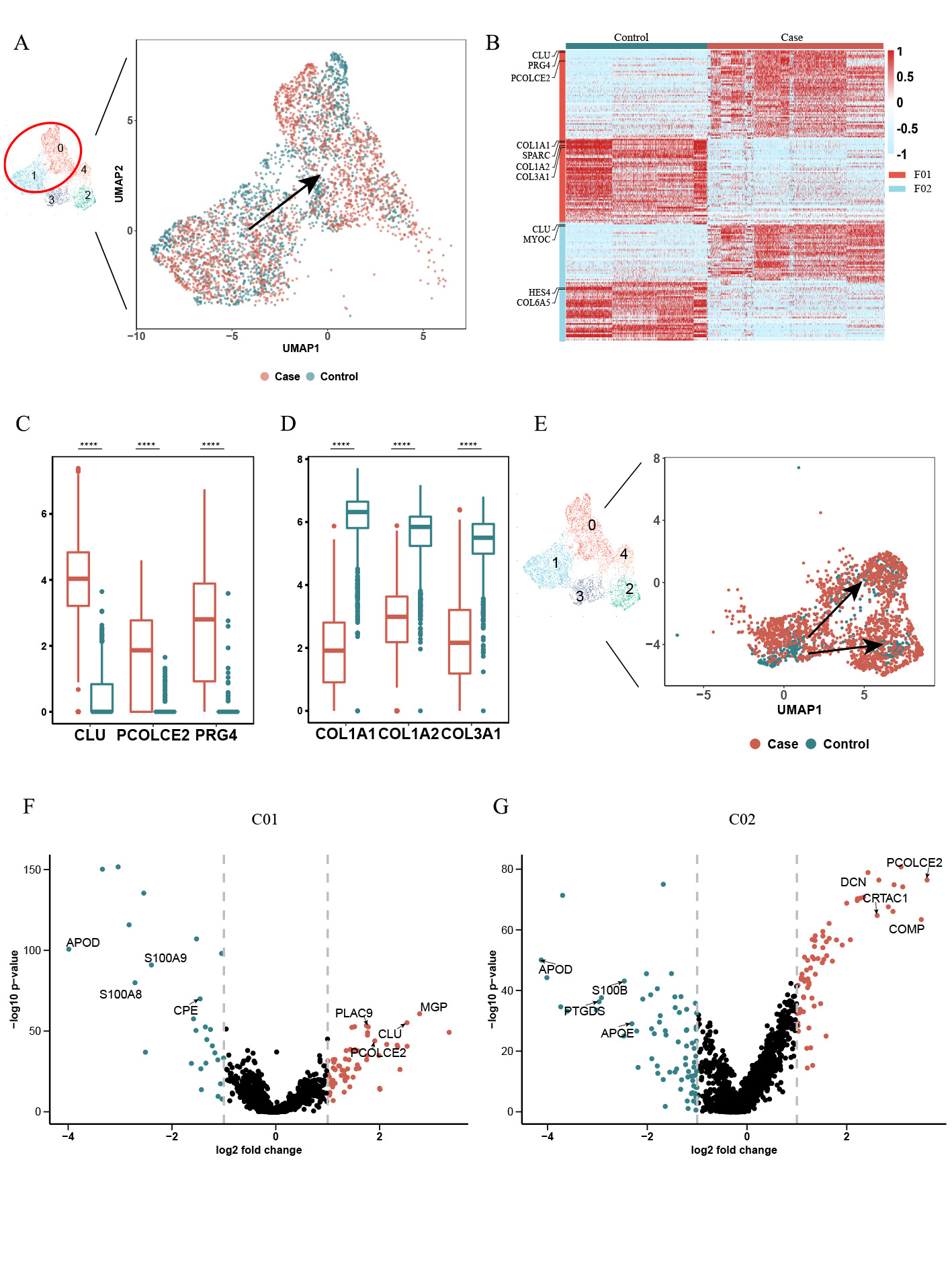
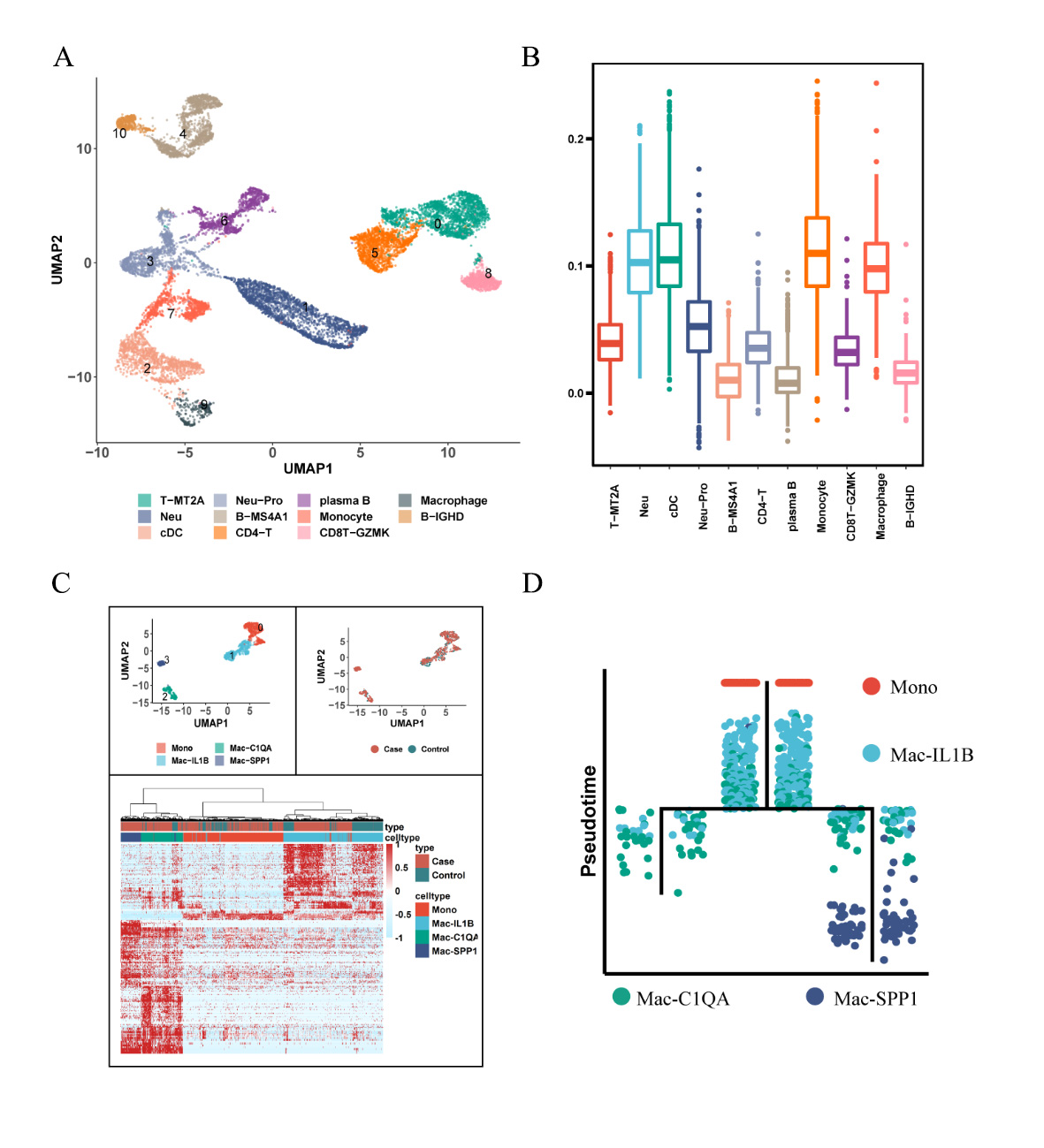
Results: After rigorous quality control, we obtained high-quality data from 32,014 cells for downstream analysis (Figure 1A). Our findings revealed distinct cell clusters comprising stromal and immune cells (Figures 1B-C). Stromal cells exhibited five cell subpopulations (Figures 1D-F), including GPX3+ tenocytes, two clusters of chondrocyte-like cells (CHAD+ and CRTAC1+), and two clusters of fibroblast-like cells (CTHRC1+ and CXCL14+). Notably, collagen degradation associated with CTHRC1+ fibroblast-like cells (Figures 2A-D) and ossification linked to CRTAC1+ chondrocyte-like cells (Figures 2E-G) were key features of ligament degeneration. Immune cells exhibited 11 cell subpopulations, including monocyte/macrophage, neutrophils, T cell, B cell, plasma B and cDC (Figure 3A). Among immune cells, myeloid cells demonstrated pathogenic activation, and degenerative ligaments showed enrichment of SPP1+ macrophages (Figures 3B-D). Further investigations unveiled the influence of ATF3, a core transcription factor, on the expression of CRTAC1/MGP/CLU in stromal cells, potentially mediating the interaction between SPP1+ macrophages and ligament degeneration (data not shown here).
Conclusion: SPP1+ macrophages contribute to the pathology of ligament degeneration through their interaction with the core transcription factor ATF3, resulting in enhanced expression of CRTAC1/MGP/CLU in stromal cells. Targeting SPP1+ macrophages and ATF3 in ligament stromal cells may hold promise for mitigating ligament degeneration.
To cite this abstract in AMA style:
Tang Y, Zhuo D, Jin L, Zhu Q, Chen Y, Wang J, Liu J. Single-cell RNA Sequencing Reveals the CRTAC1+ Population Actively Contributes to the Pathogenesis of Spinal Ligament Degeneration by Inducing Macrophage Activation [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/single-cell-rna-sequencing-reveals-the-crtac1-population-actively-contributes-to-the-pathogenesis-of-spinal-ligament-degeneration-by-inducing-macrophage-activation/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-rna-sequencing-reveals-the-crtac1-population-actively-contributes-to-the-pathogenesis-of-spinal-ligament-degeneration-by-inducing-macrophage-activation/