Session Information
Session Type: Abstract Session
Session Time: 9:15AM-9:30AM
Background/Purpose: Although antiphospholipid syndrome (APS) regularly presents with discrete thrombotic events, many patients will also acquire organ damage over time secondary to occlusive neointimal formation in small vessels. The presence of livedo reticularis is predictive of various “internal” manifestations of APS including cerebrovascular disease. Although the interplay between antiphospholipid antibodies (aPL) and endothelial cells is likely a key driver of APS vasculopathy in skin and other organs, APS microvascular endothelial cells have been studied only sparsely. Here, we employed skin biopsy and sc-RNAseq to characterize APS endothelial cells with an eye toward identifying the dysregulated pathways and cellular crosstalk that may conspire to promote APS vasculopathy over time.
Methods: We applied sc-RNAseq to skin biopsies of three primary APS patients with livedo reticularis and four healthy controls with neither APS nor livedo. Uniform manifold approximation and projection (UMAP) was used to discriminate cell types, and differential gene expression analysis was identified. Pathway enrichment was determined by Ingenuity Pathway Analysis. Ligand-receptor pairs from the Database of Interacting Proteins were used to identify potential intra- and inter-cellular communication. Immunohistochemistry (IHC) was performed to verify protein expression in APS and control skin. Control human MVECs were cultured with APS sera in pursuit of discerning transcriptional expression of upregulated genes.
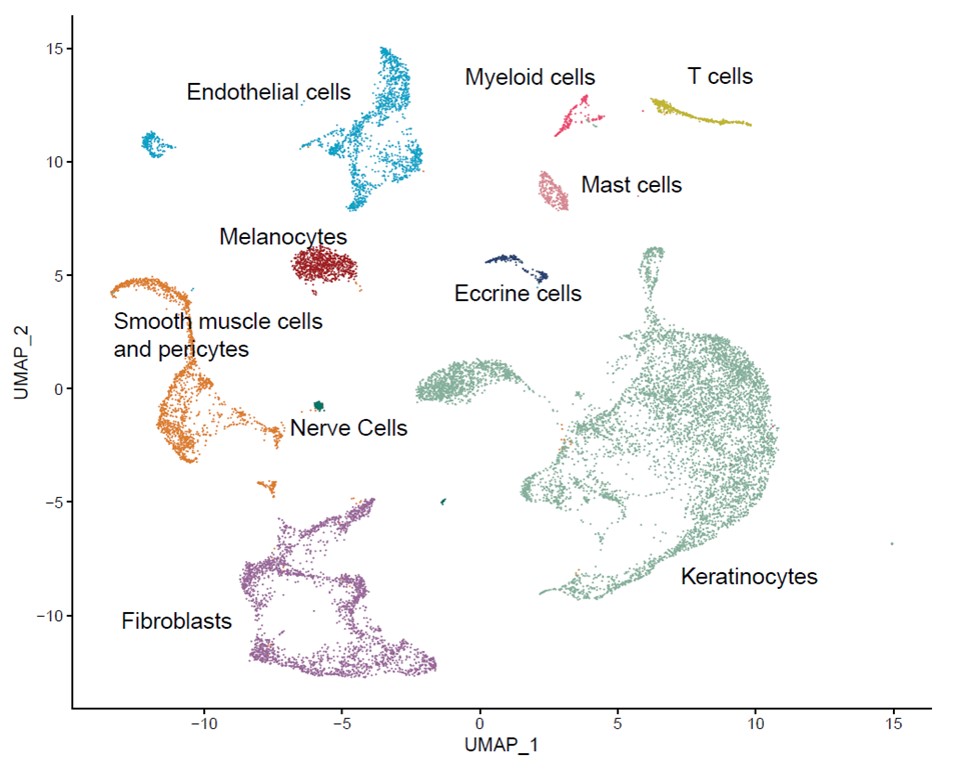
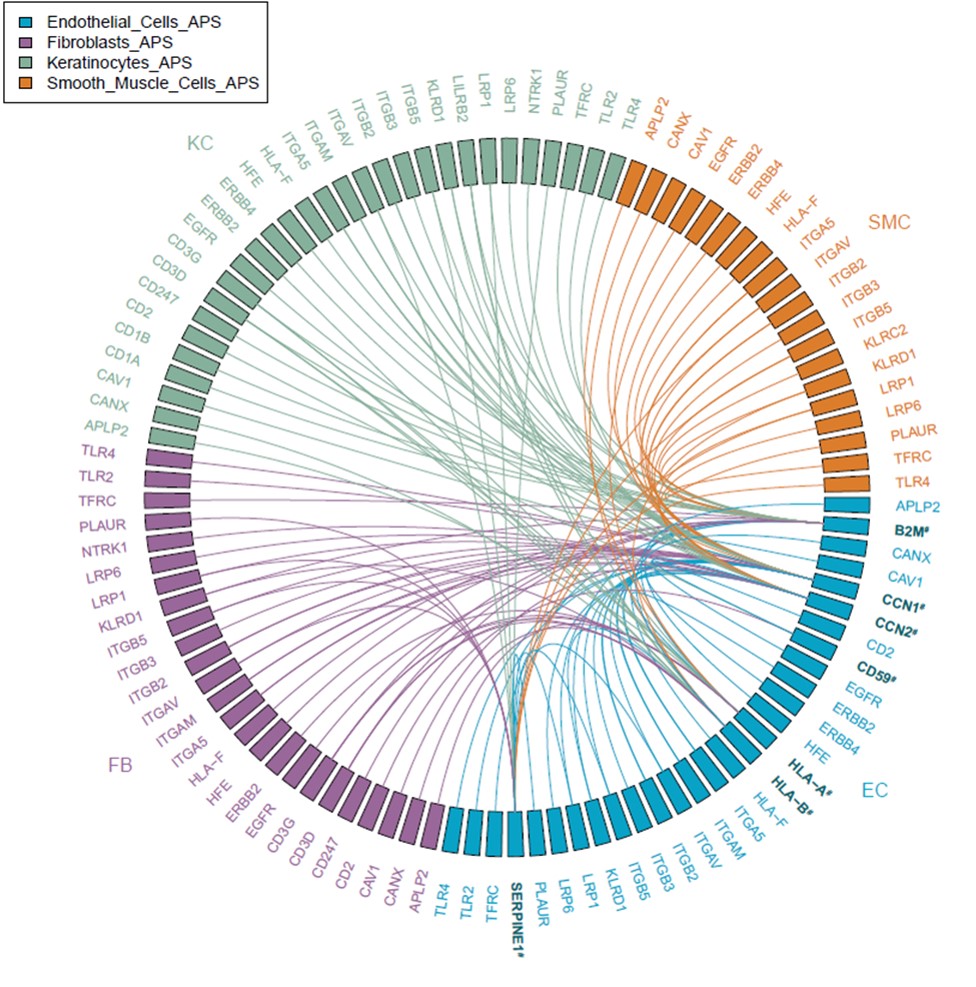
Results: We identified ten cell types in skin (Figure 1). Within the main endothelial cell population, there were four distinct subsets that we named Arterial, Capillary I, Capillary II, and Venous based on differential expression of key genes (Figure 2). We compared gene expression profiles between APS and control endothelial cells and found significant upregulation of various genes associated with cell proliferation, most notably CCN1 (CYR61), CCN2 (CTGF), and SERPINE1 (PAI-1) in APS. These changes were most pronounced in the Capillary I and Venous subsets. By pathway analysis, we found the APS endothelial cell expression profile to be enriched for Hippo-YAP1/TAZ and TGF-β pathways. Anchored by differentially-expressed ligands of APS endothelial cells, interaction analysis suggested strong communication between endothelial cells (especially ligands CCN1 and CCN2) and both pericytes/smooth muscle cells and fibroblasts (Figure 3). IHC staining confirmed higher expression of CCN1 and CCN2 in APS skin as compared with control skin. In these same biopsies, APS endothelial cells were more likely to display YAP1 nuclear translocation as compared with control endothelial cells. In vitro, culture of control MVECs with APS sera upregulated CCN1, CCN2, and SERPINE1.
Conclusion: To our knowledge, this is the first direct analysis of gene expression in APS endothelial cells and local interacting cells. These data suggest that APS endothelial cells demonstrate a pattern of ligand gene expression with potential to drive the proliferation of neighboring cells including pericytes/smooth muscle cells and fibroblasts, which may have significant relevance for the neointimal formation of APS vasculopathy.
To cite this abstract in AMA style:
Shi H, Billi A, Wasikowski R, Hoy C, Gockman K, Tsoi L, Gudjonsson J, Tsou P, Knight J. Single-Cell RNA Sequencing of APS Skin Reveals Endothelial Pathology and Cellular Interactions [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/single-cell-rna-sequencing-of-aps-skin-reveals-endothelial-pathology-and-cellular-interactions/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-rna-sequencing-of-aps-skin-reveals-endothelial-pathology-and-cellular-interactions/