Session Information
Date: Sunday, October 26, 2025
Title: (0067–0097) Rheumatoid Arthritis – Etiology and Pathogenesis Poster
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Interstitial lung disease (ILD) is one of the extra-articular manifestations of rheumatoid arthritis (RA) characterized by inflammation and/or fibrosis. Clinically relevant RA-ILD occurs in approximately 10% of RA patients and is associated with poor prognosis. Recent single-cell RNA-sequencing (scRNA-seq) analysis have revealed the effects of the cellular makeup and molecular pathways on pathological changes in several lung diseases, but little is known about RA-ILD. This study aimed to characterize immune/epithelial cells in RA-ILD through scRNA-seq analysis.
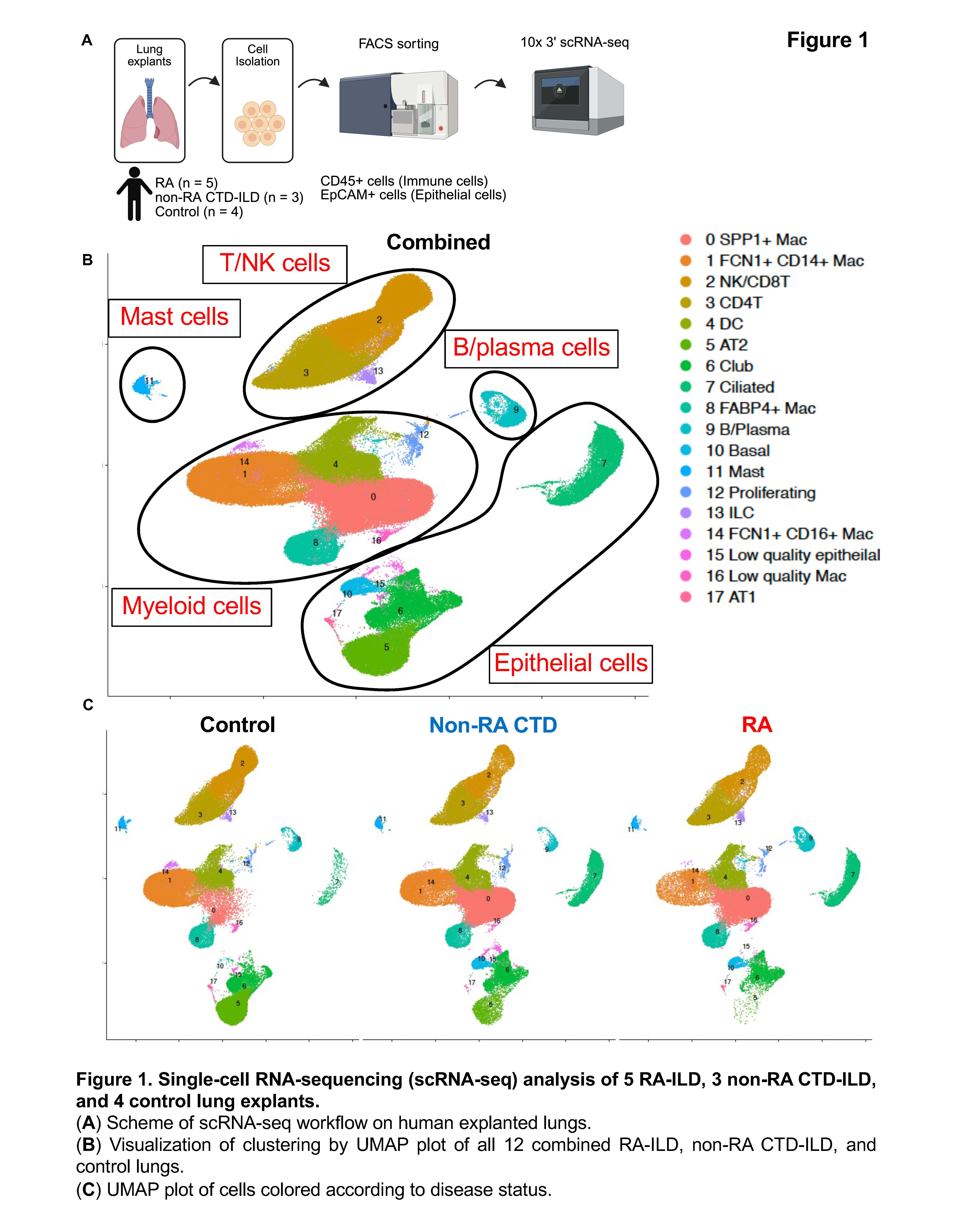
Methods: We processed explant tissues from 5 RA-ILD patients, 3 non-RA connective tissue disease (CTD)-ILD (2 with dermatomyositis (DM) and 1 mixed connective tissue disease (MCTD)), and 4 control patients. CD45+CD31-EpCAM- immune cells and CD45-CD31-EpCAM+ epithelial cells were FACS sorted and subjected to scRNA-seq. Ingenuity pathway analysis was used to perform upstream regulator analysis.
Results: We analyzed 184,814 cells after quality-control filtering and identified 18 distinct cell clusters (Figure 1). By differential abundance analysis, we found that AT2 cells were 65% lower in abundance in RA-ILD lung tissue compared with control lung tissue, and 10% lower in RA-ILD compared with non-RA CTD-ILD. We identified a reciprocal higher abundance of other epithelial cell types (basal cells, club cells, and ciliated cells). We also found fewer FCN1+ CD14+ macrophages and more SPP1+ macrophages in RA-ILD lungs. We calculated the number of differentially expressed genes (DEGs) in each cell cluster between the disease groups (RA vs control and RA vs CTD) and found that AT2 cells, club cells, FCN1+ CD14+ macrophages, FABP4+ macrophages, and SPP1+ macrophage had a higher number of DEGs (Figure 2). Upstream regulator analysis revealed that TNFa was potentially activated as an upstream molecule which drived differential changes in RA-ILD AT2 cells. T cell sub-clustering analysis revealed that peripheral T cells (Tph) are exclusively found in RA-ILD lungs (Figure 3).
Conclusion: There was heterogeneity in several key cell types, such as AT2 cells, club cells, FCN1+ CD14+ macrophages, FABP4+ macrophages, SPP1+ macrophages, and Tph cells between RA-ILD, non-RA CTD-ILD, and control lungs. An upstream regulator analysis suggested that TNF signaling might be involved in the phenotypic change of AT2 cells in RA-ILD. Our study suggests that immune/epithelial cell abnormalities underlie the pathogenesis of RA-ILD.
To cite this abstract in AMA style:
Suga K, Seng A, Yao C, Parimon T, Choi Y, Fert-Bober J, Stripp B, Giles J, Chen P, Bottini N. Single Cell RNA-seq Revealed Immune/epithelial Cell Abnormalities Underlying the Pathogenesis of Rheumatoid Arthritis-related Interstitial Lung Disease [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/single-cell-rna-seq-revealed-immune-epithelial-cell-abnormalities-underlying-the-pathogenesis-of-rheumatoid-arthritis-related-interstitial-lung-disease/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-rna-seq-revealed-immune-epithelial-cell-abnormalities-underlying-the-pathogenesis-of-rheumatoid-arthritis-related-interstitial-lung-disease/

.jpg)
.jpg)