Session Information
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Psoriatic arthritis (PsA) affects males and females equally, but differences exist in the clinical presentation and treatment response. The biological mechanisms driving these differences are unknown. We aimed to identify sex-specific serum proteins and biological pathways in male vs. female patients with PsA compared to controls.
Methods: We analyzed 100 age- and sex-matched active PsA patients and 50 age- and sex-matched healthy controls using an aptamer-based assay to measure over 7,000 serum proteins. Differential analysis was conducted for PsA males vs. PsA females and PsA vs. controls overall and by sex using the Limma package in R. Differentially expressed proteins (DEPs) were identified using a false discovery rate p < 0.05 and a fold change >1.2. We used pathDIP to identify sex-specific enriched pathways from the DEPs among male vs. female PsA and constructed a protein-pathway network using NAViGaTOR. Additionally, we applied various supervised classifiers to identify protein biomarkers contributing to the differences between PsA and controls, including logistic regression with elastic net, linear discriminant analysis, support vector machine, and random forest using the caret package. Using a random forest and variable importance analysis, we report the protein biomarkers that contributed the most to discriminating PsA from controls for male and female patients separately.
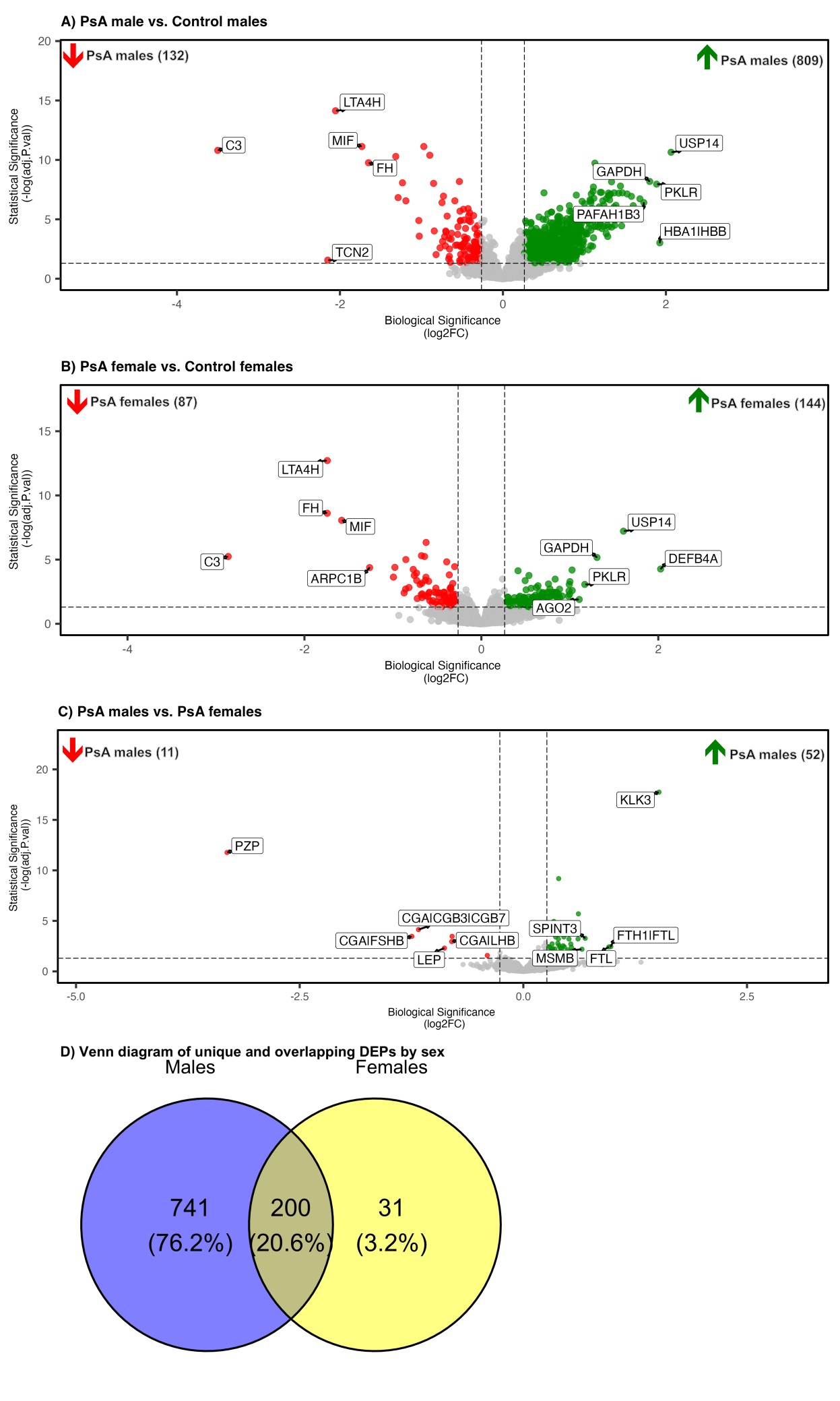
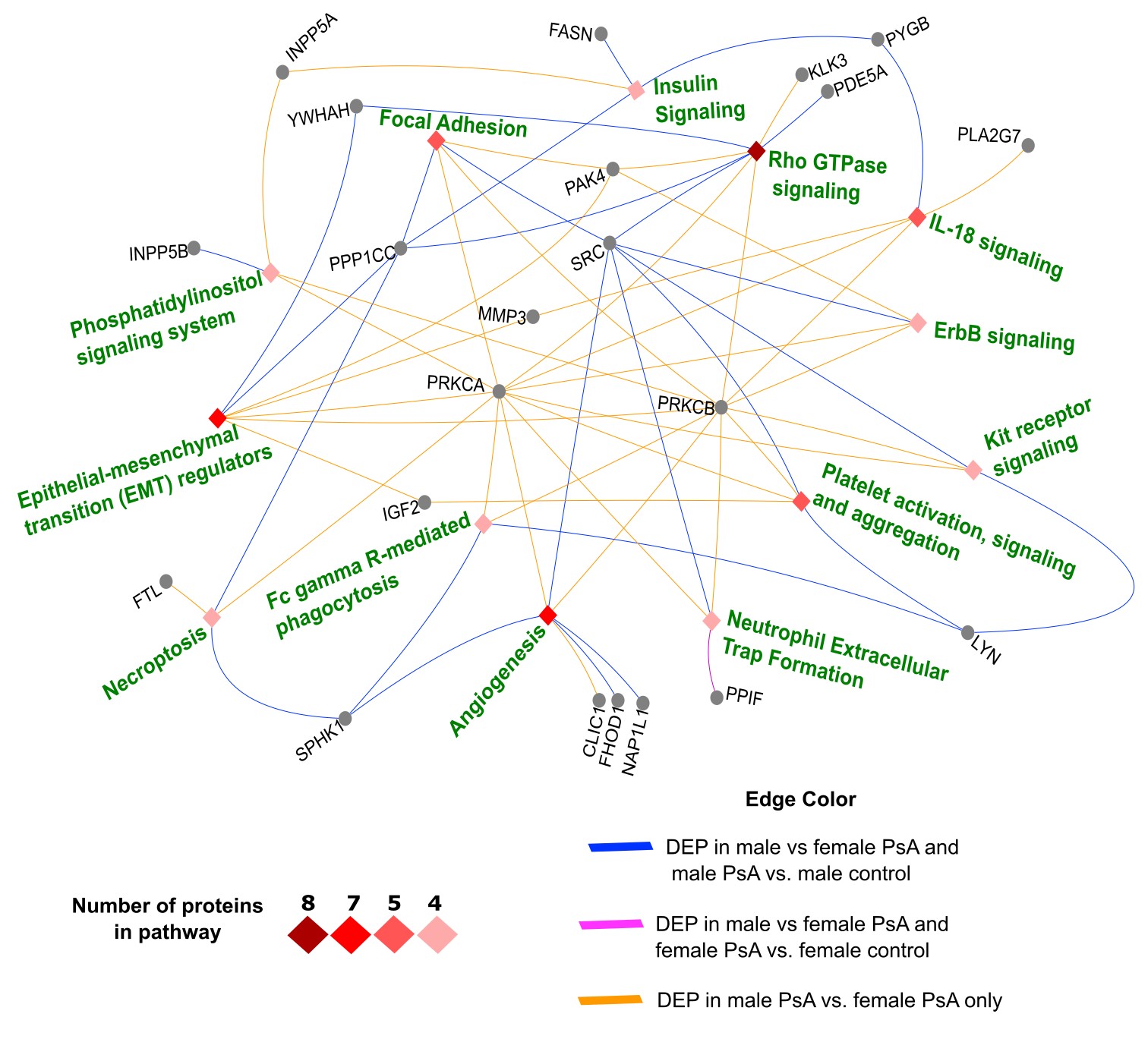
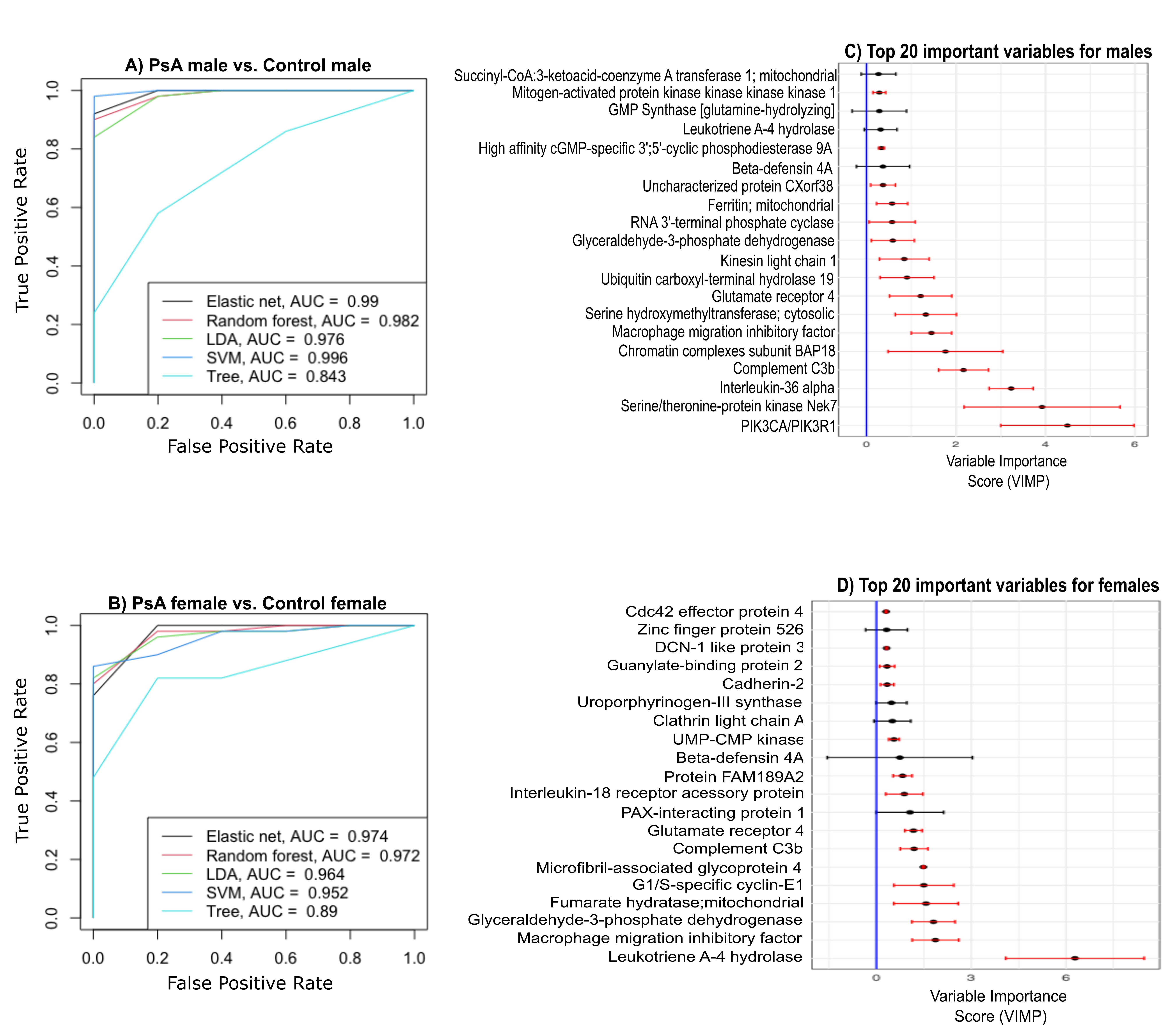
Results: The differential analysis revealed a significantly higher number of sex-specific, unique DEPs in PsA males vs. controls (741) compared to PsA females vs. controls (31), and 200 proteins were shared in males and females with PsA compared to controls (Figure 1A-D). Several sex-specific pathways were identified among male vs. female PsA patients, such as Rho GTPase signaling, angiogenesis, focal adhesion, IL-18 signaling, neutrophil extracellular trap (NET) formation, Fc gamma R-mediated phagocytosis, platelet function, ErbB signaling, kit receptor signaling, phosphatidylinositol signaling, and epithelial-mesenchymal transition regulators (Figure 2). More proteins belonging to these pathways that were both sex- and PsA-specific proteins were found in males (10 proteins, e.g., SRC, LYN, PDE5A) than in females (1 protein, PPIF). Lastly, multi-protein biomarker classifiers for PsA status vs. control performed well among males and females (AUC > 0.9 for most classifiers; Figure 3). Random forest analysis identified several mutual proteins to males and females with PsA (e.g., macrophage migration inhibitory factor, C3b) and others that were male-specific (e.g., PI3KCA/PIK3R1, Nek7, IL-36 alpha), and female-specific (e.g., G1/S-specific cyclin E1, mitochondrial fumarate hydratase).
Conclusion: In this untargeted multi-protein analysis, we provide evidence of sex-related biological differences in PsA. More unique proteins and pathways were discovered in male than female PsA patients. These sex-specific pathways are related to immune cell function (e.g., NETosis, phagocytosis), cytokine signaling, and intracellular signaling (e.g., Rho GTPase signaling). These findings offer potential new targets for future sex-based research in PsA.
To cite this abstract in AMA style:
Dang S, Li X, Diao L, Wither J, Jurisica I, Chandran V, Eder L. Sex Differences in Serum Proteomic Profiles of Males and Females with Psoriatic Arthritis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/sex-differences-in-serum-proteomic-profiles-of-males-and-females-with-psoriatic-arthritis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/sex-differences-in-serum-proteomic-profiles-of-males-and-females-with-psoriatic-arthritis/