Session Information
Date: Monday, November 9, 2020
Title: SLE – Diagnosis, Manifestations, & Outcomes Poster III: Bench to Bedside
Session Type: Poster Session D
Session Time: 9:00AM-11:00AM
Background/Purpose: SLE is a heterogeneous disease, where a better understanding of molecular differences between patients is needed in order to direct therapy. Existing approaches generally examine mRNA expression, whereas therapeutic targets are mostly soluble or cellular proteins. We aimed to evaluate whether a serum cytokine proteomic profile of SLE patients analysed using an unbiased approach would yield clinically meaningful results.
Methods: Demographic and clinical data of SLE patients (ACR criteria) including disease activity (SLEDAI2K) and organ damage (SLICC Damage Index (SDI)), and matching serum samples, were collected prospectively. A wide-angled serum cytokine proteomics analysis was conducted using Quantibody, Luminex, and ELISA platforms. To reduce heterogeneous data complexity and remove redundant variables, unsupervised feature selection methods were applied. For clustering, machine learning approaches were used, and optimal cluster number confirmed by consensus clustering.
Results: 198 SLE patients (median [IQR] age 46.7 [36.7, 56.3] years, 88.4% female, median SLEDAI2K 4 [2, 6], 52.5% with organ damage) and 37 sex/ethnicity-matched healthy subjects were recruited. 211 serum analytes were measured. Using unsupervised feature selection methods, we identified a reduced set of 10/211 analytes (MCP-1, GH, CTACK, HCC-1, CD14, ErbB3, E-Selectin, Trappin-2, Cathepsin S and IL-18). The dataset was then clustered, according to the analytes that remained after feature selection, using unsupervised machine learning approaches. k-means analysis produced a distinct result in terms of separability of two clusters that associated only in SLE (Figure 1), and was chosen as the final clustering algorithm. We analysed clinical parameters in patients categorised by these biomarker clusters, and found the clusters differed in clinical characteristics including organ damage (P = 0.04), proportion of patients with high ESR (P = 0.03), and SLEDAI2K (P = 0.08). Using multivariable linear regression models, cross-sectional associations with patient clinical characteristics were found for 8/10 analytes. We next assessed associations of baseline analytes with longitudinal disease outcomes, using multivariable logistic regression models. Trappin-2 and IL-18 were significantly associated with damage accrual. IL-18 and CTACK had positive and negative associations respectively with lupus low disease activity state (LLDAS) attainment, and converse associations with indices of active disease over time.
Conclusion: Unsupervised analytics of wide-angle serum cytokine profiles in SLE yielded a tractable reduced analyte set, which in turn revealed two biologically distinct clusters of patients who had significant differences in clinical profile, individual analytes from which were associated with longitudinal outcomes. These findings indicate the potential for biological subsets of SLE to be based on serum cytokine profiling.
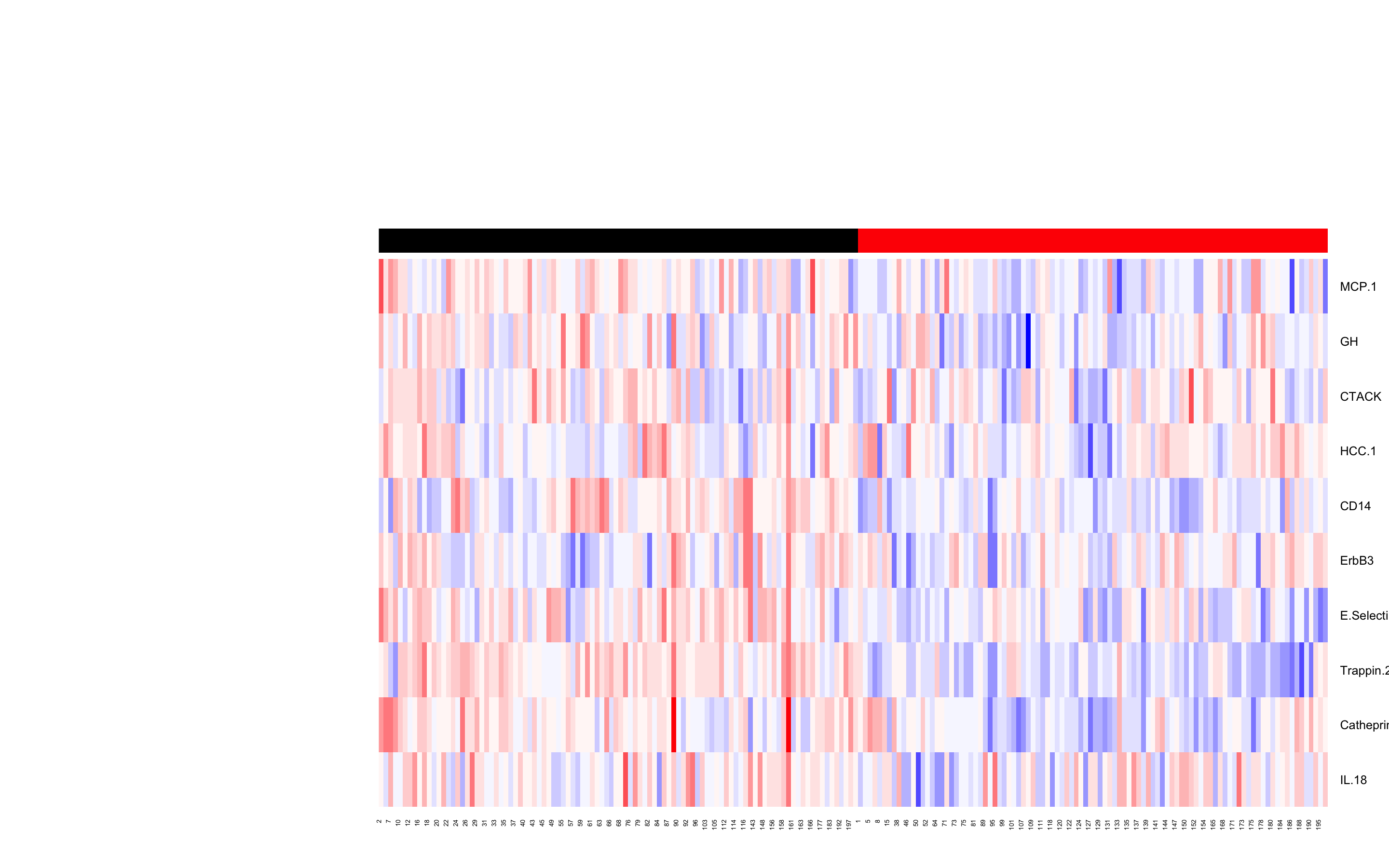 Figure 1. Heat map of 10 analytes, selected using unsupervised feature selection on 211 analytes measured in healthy subjects and SLE patients and clustered using k-means techniques, in SLE patients (n=198). Two clusters (indicated by red or black on top row) were identified in SLE.
Figure 1. Heat map of 10 analytes, selected using unsupervised feature selection on 211 analytes measured in healthy subjects and SLE patients and clustered using k-means techniques, in SLE patients (n=198). Two clusters (indicated by red or black on top row) were identified in SLE.
To cite this abstract in AMA style:
Vincent F, Ong J, Hoi A, Boyd S, Nim H, Morand E. Serum Cytokine Profiling in Systemic Lupus Erythematosus, Analysed Using Unsupervised Machine Learning, Reveals Clinically Relevant Clusters [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/serum-cytokine-profiling-in-systemic-lupus-erythematosus-analysed-using-unsupervised-machine-learning-reveals-clinically-relevant-clusters/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/serum-cytokine-profiling-in-systemic-lupus-erythematosus-analysed-using-unsupervised-machine-learning-reveals-clinically-relevant-clusters/
